Rice transcriptional repressor OsTIE1 controls anther dehiscence and male sterility by regulating JA biosynthesis
- PMID: 38299434
- PMCID: PMC11062430
- DOI: 10.1093/plcell/koae028
Rice transcriptional repressor OsTIE1 controls anther dehiscence and male sterility by regulating JA biosynthesis
Abstract
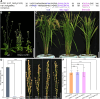
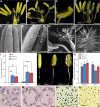
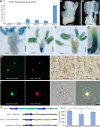
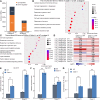
Proper anther dehiscence is essential for successful pollination and reproduction in angiosperms, and jasmonic acid (JA) is crucial for the process. However, the mechanisms underlying the tight regulation of JA biosynthesis during anther development remain largely unknown. Here, we demonstrate that the rice (Oryza sativa L.) ethylene-response factor-associated amphiphilic repression (EAR) motif-containing protein TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTORS (TCP) INTERACTOR CONTAINING EAR MOTIF PROTEIN1 (OsTIE1) tightly regulates JA biosynthesis by repressing TCP transcription factor OsTCP1/PCF5 during anther development. The loss of OsTIE1 function in Ostie1 mutants causes male sterility. The Ostie1 mutants display inviable pollen, early stamen filament elongation, and precocious anther dehiscence. In addition, JA biosynthesis is activated earlier and JA abundance is precociously increased in Ostie1 anthers. OsTIE1 is expressed during anther development, and OsTIE1 is localized in nuclei and has transcriptional repression activity. OsTIE1 directly interacts with OsTCP1, and overexpression of OsTCP1 caused early anther dehiscence resembling that of Ostie1. JA biosynthesis genes including rice LIPOXYGENASE are regulated by the OsTIE1-OsTCP1 complex. Our findings reveal that the OsTIE1-OsTCP1 module plays a critical role in anther development by finely tuning JA biosynthesis and provide a foundation for the generation of male sterile plants for hybrid seed production.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures



References
-
- Agrawal GK, Jwa N-S, Agrawal SK, Tamogami S, Iwahashi H, Rakwal R. Cloning of novel rice allene oxide cyclase (OsAOC): mRNA expression and comparative analysis with allene oxide synthase (OsAOS) gene provides insight into the transcriptional regulation of octadecanoid pathway biosynthetic genes in rice. Plant Sci. 2003:164(6):979–992. 10.1016/S0168-9452(03)00082-7 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

