Single-cell ATAC sequencing identifies sleepy macrophages during reciprocity of cytokines in L. major infection
- PMID: 38299832
- PMCID: PMC10913457
- DOI: 10.1128/spectrum.03478-23
Single-cell ATAC sequencing identifies sleepy macrophages during reciprocity of cytokines in L. major infection
Abstract
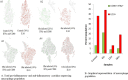
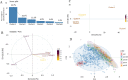
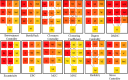
The hallmark characteristic of macrophages lies in their inherent plasticity, allowing them to adapt to dynamic microenvironments. Leishmania strategically modulates the phenotypic plasticity of macrophages, creating a favorable environment for intracellular survival and persistent infection through regulatory cytokine such as interleukin (IL)-10. Nevertheless, these effector cells can counteract infection by modulating crucial cytokines like IL-12 and key components involved in its production. Using sophisticated tool of single-cell assay for transposase accessible chromatin (ATAC) sequencing, we systematically examined the regulatory axis of IL-10 and IL-12 in a time-dependent manner during Leishmania major infection in macrophages Our analysis revealed the cellular heterogeneity post-infection with the regulators of IL-10 and IL-12, unveiling a reciprocal relationship between these cytokines. Notably, our significant findings highlighted the presence of sleepy macrophages and their pivotal role in mediating reciprocity between IL-10 and IL-12. To summarize, the roles of cytokine expression, transcription factors, cell cycle, and epigenetics of host cell machinery were vital in identification of sleepy macrophages, which is a transient state where transcription factors controlled the epigenetic remodeling and expression of genes involved in pro-inflammatory cytokine expression and recruitment of immune cells.IMPORTANCELeishmaniasis is an endemic affecting 99 countries and territories globally, as outlined in the 2022 World Health Organization report. The disease's severity is compounded by compromised host immune systems, emphasizing the pivotal role of the interplay between parasite and host immune factors in disease regulation. In instances of cutaneous leishmaniasis induced by L. major, macrophages function as sentinel cells. Our findings indicate that the plasticity and phenotype of macrophages can be modulated to express a cytokine profile involving IL-10 and IL-12, mediated by the regulation of transcription factors and their target genes post-L. major infection in macrophages. Employing sophisticated methodologies such as single-cell ATAC sequencing and computational genomics, we have identified a distinctive subset of macrophages termed "sleepy macrophages." These macrophages exhibit downregulated housekeeping genes while expressing a unique set of variable features. This data set constitutes a valuable resource for comprehending the intricate host-parasite interplay during L. major infection.
Keywords: parasite; single-cell ATAC sequencing; sleepy macrophages; systems biology; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

