Swine influenza A virus isolates containing the pandemic H1N1 origin matrix gene elicit greater disease in the murine model
- PMID: 38299860
- PMCID: PMC10913740
- DOI: 10.1128/spectrum.03386-23
Swine influenza A virus isolates containing the pandemic H1N1 origin matrix gene elicit greater disease in the murine model
Abstract
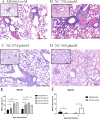
Since the 1990s, endemic North American swine influenza A viruses (swFLUAVs) contained an internal gene segment constellation, the triple reassortment internal gene (TRIG) cassette. In 2009, the H1N1 pandemic (pdmH1N1) virus spilled back into swine but did not become endemic. However, the pdmH1N1 contributed the matrix gene (pdmM) to the swFLUAVs circulating in the pig population, which replaced the classical swine matrix gene (swM) found in the TRIG cassette, suggesting the pdmM has a fitness benefit. Others have shown that swFLUAVs containing the pdmM have greater transmission efficiency compared to viruses containing the swM gene segment. We hypothesized that the matrix (M) gene could also affect disease and utilized two infection models, resistant BALB/c and susceptible DBA/2 mice, to assess pathogenicity. We infected BALB/c and DBA/2 mice with H1 and H3 swFLUAVs containing the swM or pdmM and measured lung virus titers, morbidity, mortality, and lung histopathology. H1 influenza strains containing the pdmM gene caused greater morbidity and mortality in resistant and susceptible murine strains, while H3 swFLUAVs caused no clinical disease. However, both H1 and H3 swFLUAVs containing the pdmM replicated to higher viral titers in the lungs and pdmM containing H1 viruses induced greater histological changes compared to swM H1 viruses. While the surface glycoproteins and other gene segments may contribute to swFLUAV pathogenicity in mice, these data suggest that the origin of the matrix gene also contributes to pathogenicity of swFLUAV in mice, although we must be cautious in translating these conclusions to their natural host, swine.
Importance: The 2009 pandemic H1N1 virus rapidly spilled back into North American swine, reassorting with the already genetically diverse swFLUAVs. Notably, the M gene segment quickly replaced the classical M gene segment, suggesting a fitness benefit. Here, using two murine models of infection, we demonstrate that swFLUAV isolates containing the pandemic H1N1 origin M gene caused increased disease compared to isolates containing the classical swine M gene. These results suggest that, in addition to other influenza virus gene segments, the swFLUAV M gene segment contributes to pathogenesis in mammals.
Keywords: influenza virus; matrix gene; mouse model; pandemic influenza; pathogenesis; swine influenza.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Virologic Differences Do Not Fully Explain the Diversification of Swine Influenza Viruses in the United States.J Virol. 2016 Oct 28;90(22):10074-10082. doi: 10.1128/JVI.01218-16. Print 2016 Nov 15. J Virol. 2016. PMID: 27581984 Free PMC article.
-
Comparative In Vitro and In Vivo Analysis of H1N1 and H1N2 Variant Influenza Viruses Isolated from Humans between 2011 and 2016.J Virol. 2018 Oct 29;92(22):e01444-18. doi: 10.1128/JVI.01444-18. Print 2018 Nov 15. J Virol. 2018. PMID: 30158292 Free PMC article.
-
Pathogenesis of pandemic influenza A (H1N1) and triple-reassortant swine influenza A (H1) viruses in mice.J Virol. 2010 May;84(9):4194-203. doi: 10.1128/JVI.02742-09. Epub 2010 Feb 24. J Virol. 2010. PMID: 20181710 Free PMC article.
-
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese.
-
Diversity of influenza viruses in swine and the emergence of a novel human pandemic influenza A (H1N1).Influenza Other Respir Viruses. 2009 Sep;3(5):207-13. doi: 10.1111/j.1750-2659.2009.00096.x. Influenza Other Respir Viruses. 2009. PMID: 19768134 Free PMC article. Review.
Cited by
-
Establishment of Swine Primary Nasal, Tracheal, and Bronchial Epithelial Cell Culture Models for the Study of Influenza Virus Infection.J Virol Methods. 2024 Jun;327:114943. doi: 10.1016/j.jviromet.2024.114943. Epub 2024 Apr 26. J Virol Methods. 2024. PMID: 38679164 Free PMC article.
-
Pandemic Risk Assessment for Swine Influenza A Virus in Comparative In Vitro and In Vivo Models.Viruses. 2024 Mar 31;16(4):548. doi: 10.3390/v16040548. Viruses. 2024. PMID: 38675891 Free PMC article.
References
-
- Shaw ML, Palese P. 2013. Orthomyxoviridae, p 2 volumes. In Knipe DM, Howley PM (ed), Fields virology, 6th ed. Wolters Kluwer/Lippincott Williams & Wilkins Health, Philadelphia, PA.
-
- Organization WH. 2014. Influenza (seasonal) fact sheet N211
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

