Single-cell Multiomics Analysis of Myelodysplastic Syndromes and Clinical Response to Hypomethylating Therapy
- PMID: 38300528
- PMCID: PMC10860538
- DOI: 10.1158/2767-9764.CRC-23-0389
Single-cell Multiomics Analysis of Myelodysplastic Syndromes and Clinical Response to Hypomethylating Therapy
Abstract
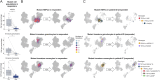
Alterations in epigenetic marks, such as DNA methylation, represent a hallmark of cancer that has been successfully exploited for therapy in myeloid malignancies. Hypomethylating agents (HMA), such as azacitidine, have become standard-of-care therapy to treat myelodysplastic syndromes (MDS), myeloid neoplasms that can evolve into acute myeloid leukemia. However, our capacity to identify who will respond to HMAs, and the duration of response, remains limited. To shed light on this question, we have leveraged the unprecedented analytic power of single-cell technologies to simultaneously map the genome and immunoproteome of MDS samples throughout clinical evolution. We were able to chart the architecture and evolution of molecular clones in precious paired bone marrow MDS samples at diagnosis and posttreatment to show that a combined imbalance of specific cell lineages with diverse mutational profiles is associated with the clinical response of patients with MDS to hypomethylating therapy.
Significance: MDS are myeloid clonal hemopathies with a low 5-year survival rate, and approximately half of the cases do not respond to standard HMA therapy. Our innovative single-cell multiomics approach offers valuable biological insights and potential biomarkers associated with the demethylating agent efficacy. It also identifies vulnerabilities that can be targeted using personalized combinations of small drugs and antibodies.
© 2024 The Authors; Published by the American Association for Cancer Research.
Figures





References
-
- Tefferi A, Vardiman JW. Myelodysplastic syndromes. N Engl J Med 2009;361:1872–85. - PubMed
-
- Sekeres MA, Taylor J. Diagnosis and treatment of myelodysplastic syndromes: a review. JAMA 2022;328:872–80. - PubMed
-
- Greenberg PL, Stone RM, Al-Kali A, Bennett JM, Borate U, Brunner AM, et al. . NCCN guidelines® insights: myelodysplastic syndromes, version 3.2022. J Natl Compr Canc Netw 2022;20:106–17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 2017 SGR1080/Government of Catalonia | Departament d'Empresa i Coneixement, Generalitat de Catalunya (Ministry of Business and Knowledge, Government of Catalonia)
- 2021 SGR01494/Government of Catalonia | Departament d'Empresa i Coneixement, Generalitat de Catalunya (Ministry of Business and Knowledge, Government of Catalonia)
- 2017 SGR288 2021/Government of Catalonia | Departament d'Empresa i Coneixement, Generalitat de Catalunya (Ministry of Business and Knowledge, Government of Catalonia)
- 2021 SGR00560/Government of Catalonia | Departament d'Empresa i Coneixement, Generalitat de Catalunya (Ministry of Business and Knowledge, Government of Catalonia)
- RTI2018-094049-BI00/Ministerio de Ciencia e Innovación (MCIN)
- PID2021-125282OB-I00/Ministerio de Ciencia e Innovación (MCIN)
- CEL007/Fundación Cellex (Cellex Foundation)
- LCF/PR/HR22/00732/Fundación Bancaria Caixa d'Estalvis i Pensions de Barcelona ("la Caixa" Foundation)
- PI/17/00575/Ministerio de Ciencia e Innovación (MCIN)
- PI 20/00531/Ministerio de Ciencia e Innovación (MCIN)
- AC 18/000002/Fundación Científica Asociación Española Contra el Cáncer (AECC)
- FJC2020-044658-I/Ministerio de Ciencia e Innovación (MCIN)
- H2020-MSCA-IF-2019-896403/EC | Horizon Europe | Excellent Science | HORIZON EUROPE Marie Sklodowska-Curie Actions (MSCA)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

