HER2 heterogeneity and treatment response-associated profiles in HER2-positive breast cancer in the NCT02326974 clinical trial
- PMID: 38300710
- PMCID: PMC10977978
- DOI: 10.1172/JCI176454
HER2 heterogeneity and treatment response-associated profiles in HER2-positive breast cancer in the NCT02326974 clinical trial
Abstract
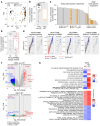
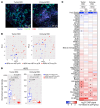
BACKGROUNDHER2-targeting therapies have great efficacy in HER2-positive breast cancer, but resistance, in part due to HER2 heterogeneity (HET), is a significant clinical challenge. We previously described that in a phase II neoadjuvant trastuzumab emtansine (T-DM1) and pertuzumab (P) clinical trial in early-stage HER2-positive breast cancer, none of the patients with HER2-HET tumors had pathologic complete response (pCR).METHODSTo investigate cellular and molecular differences among tumors according to HER2 heterogeneity and pCR, we performed RNA sequencing and ERBB2 FISH of 285 pretreatment and posttreatment tumors from 129 patients in this T-DM1+P neoadjuvant trial. A subset of cases was also subject to NanoString spatial digital profiling.RESULTSPretreatment tumors from patients with pCR had the highest level of ERBB2 mRNA and ERBB signaling. HER2 heterogeneity was associated with no pCR, basal-like features, and low ERBB2 expression yet high ERBB signaling sustained by activation of downstream pathway components. Residual tumors showed decreased HER2 protein levels and ERBB2 copy number heterogeneity and increased PI3K pathway enrichment and luminal features. HET tumors showed minimal treatment-induced transcriptomic changes compared with non-HET tumors. Immune infiltration correlated with pCR and HER2-HET status.CONCLUSIONResistance mechanisms in HET and non-HET tumors are distinct. HER2-targeting antibodies have limited efficacy in HET tumors. Our results support the stratification of patients based on HET status and the use of agents that target downstream components of the ERBB signaling pathway in patients with HET tumors.TRIAL REGISTRATIONClinicalTrials.gov NCT02326974.FUNDINGThis study was funded by Roche and the National Cancer Institute.
Keywords: Breast cancer; Oncology.
Figures





References
-
- Emens LA, et al. Overall survival (OS) in KATE2, a phase II study of programmed death ligand 1 (PD-L1) inhibitor atezolizumab (atezo)+trastuzumab emtansine (T-DM1) vs placebo (pbo)+T-DM1 in previously treated HER2+ advanced breast cancer (BC) Ann Oncol. 2019;30(suppl 5):v104. doi: 10.1093/annonc/mdz242. - DOI
-
- Filho OM, et al. Impact of HER2 heterogeneity on treatment response of early-stage HER2-positive breast cancer: phase II neoadjuvant clinical trial of T-DM1 combined with pertuzumab. Cancer Discov. 2021;11(10):2474–2487. doi: 10.1158/2159-8290.CD-20-1557. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

