FlyBase: updates to the Drosophila genes and genomes database
- PMID: 38301657
- PMCID: PMC11075543
- DOI: 10.1093/genetics/iyad211
FlyBase: updates to the Drosophila genes and genomes database
Abstract
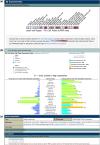
FlyBase (flybase.org) is a model organism database and knowledge base about Drosophila melanogaster, commonly known as the fruit fly. Researchers from around the world rely on the genetic, genomic, and functional information available in FlyBase, as well as its tools to view and interrogate these data. In this article, we describe the latest developments and updates to FlyBase. These include the introduction of single-cell RNA sequencing data, improved content and display of functional information, updated orthology pipelines, new chemical reports, and enhancements to our outreach resources.
Keywords: Drosophila; FlyBase; model organism database.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest: The author(s) declare no conflicts of interest.
Figures


References
-
- Alliance of Genome Resources Consortium; Agapite J, Albou L-P, Aleksander SA, Alexander M, Anagnostopoulos AV, Antonazzo G, Argasinska J, Arnaboldi V, Attrill H, et al. 2022. Harmonizing model organism data in the alliance of genome resources. Genetics. 220(4):iyac022. doi: 10.1093/genetics/iyac022. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

