Miscarriage risk assessment: a bioinformatic approach to identifying candidate lethal genes and variants
- PMID: 38302665
- PMCID: PMC10881709
- DOI: 10.1007/s00439-023-02637-y
Miscarriage risk assessment: a bioinformatic approach to identifying candidate lethal genes and variants
Abstract
Purpose: Miscarriage, often resulting from a variety of genetic factors, is a common pregnancy outcome. Preconception genetic carrier screening (PGCS) identifies at-risk partners for newborn genetic disorders; however, PGCS panels currently lack miscarriage-related genes. In this study, we evaluated the potential impact of both known and candidate genes on prenatal lethality and the effectiveness of PGCS in diverse populations.
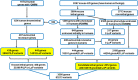
Methods: We analyzed 125,748 human exome sequences and mouse and human gene function databases. Our goals were to identify genes crucial for human fetal survival (lethal genes), to find variants not present in a homozygous state in healthy humans, and to estimate carrier rates of known and candidate lethal genes in various populations and ethnic groups.
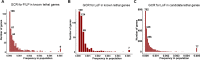
Results: This study identified 138 genes in which heterozygous lethal variants are present in the general population with a frequency of 0.5% or greater. Screening for these 138 genes could identify 4.6% (in the Finnish population) to 39.8% (in the East Asian population) of couples at risk of miscarriage. This explains the cause of pregnancy loss in approximately 1.1-10% of cases affected by biallelic lethal variants.
Conclusion: This study has identified a set of genes and variants potentially associated with lethality across different ethnic backgrounds. The variation of these genes across ethnic groups underscores the need for a comprehensive, pan-ethnic PGCS panel that includes genes related to miscarriage.
© 2024. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures



Update of
-
Preconception Genetic Carrier Screening for Miscarriage Risk Assessment: A Bioinformatic Approach to Identifying Candidate Lethal Genes and Variants.medRxiv [Preprint]. 2023 Jun 3:2023.05.25.23290518. doi: 10.1101/2023.05.25.23290518. medRxiv. 2023. Update in: Hum Genet. 2024 Feb;143(2):185-195. doi: 10.1007/s00439-023-02637-y. PMID: 37398382 Free PMC article. Updated. Preprint.
References
-
- Dixon PH, Sambrotta M, Chambers J, Taylor-Harris P, Syngelaki A, Nicolaides K, Knisely AS, Thompson RJ, Williamson C. An expanded role for heterozygous mutations of ABCB4, ABCB11, ATP8B1, ABCC2 and TJP2 in intrahepatic cholestasis of pregnancy. Sci Rep. 2017;7:11823. doi: 10.1038/s41598-017-11626-x. - DOI - PMC - PubMed
-
- Eichstadt S, Tang JY, Solis DC, Siprashvili Z, Marinkovich MP, Whitehead N, Schu M, Fang F, Erickson SW, Ritchey ME, et al. From clinical phenotype to genotypic modelling: incidence and prevalence of Recessive Dystrophic Epidermolysis Bullosa (RDEB) Clin Cosmet Investig Dermatol. 2019;12:933–942. doi: 10.2147/CCID.S232547. - DOI - PMC - PubMed
Web resources
-
- International Mouse Phenotyping Consortium http://www.mousephenotype.org/
-
- MGI-Mouse Genome Informatics http://www.informatics.jax.org/
-
- OMIM http://www.omim.org/
-
- PANTHER http://www.pantherdb.org/
-
- The Genome Aggregation Database (gnomAD) http://www.gnomad-sg.org/
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

