Modulating the skin mycobiome-bacteriome and treating seborrheic dermatitis with a probiotic-enriched oily suspension
- PMID: 38302693
- PMCID: PMC10834955
- DOI: 10.1038/s41598-024-53016-0
Modulating the skin mycobiome-bacteriome and treating seborrheic dermatitis with a probiotic-enriched oily suspension
Abstract
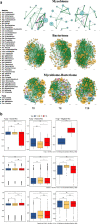
Seborrheic dermatitis (SD) affects 2-5% of the global population, with imbalances in the skin microbiome implicated in its development. This study assessed the impact of an oily suspension containing Lactobacillus crispatus P17631 and Lacticaseibacillus paracasei I1688 (termed EUTOPLAC) on SD symptoms and the skin mycobiome-bacteriome modulation. 25 SD patients were treated with EUTOPLAC for a week. Symptom severity and skin mycobiome-bacteriome changes were measured at the start of the treatment (T0), after seven days (T8), and three weeks post-treatment (T28). Results indicated symptom improvement post-EUTOPLAC, with notable reductions in the Malassezia genus. Concurrently, bacterial shifts were observed, including a decrease in Staphylococcus and an increase in Lactobacillus and Lacticaseibacillus. Network analysis highlighted post-EUTOPLAC instability in fungal and bacterial interactions, with increased negative correlations between Malassezia and Lactobacillus and Lacticaseibacillus genera. The study suggests EUTOPLAC's potential as a targeted SD treatment, reducing symptoms and modulating the mycobiome-bacteriome composition.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Dessinioti, C. & Katsambas, A. Seborrheic dermatitis: etiology, risk factors, and treatments: facts and controversies. Clin. Dermatol. 31, 343–351; 10.1016/j.clindermatol.2013.01.001 (2013). - PubMed
-
- Tao, R., Li, R. & Wang, R. Skin microbiome alterations in seborrheic dermatitis and dandruff: A systematic review. Exp. Dermatol.30, 1546–1553; 10.1111/exd.14450 (2021). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

