Transcriptomic and photosynthetic analyses of Synechocystis sp. PCC6803 and Chlorogloeopsis fritschii sp. PCC6912 exposed to an M-dwarf spectrum under an anoxic atmosphere
- PMID: 38304456
- PMCID: PMC10830646
- DOI: 10.3389/fpls.2023.1322052
Transcriptomic and photosynthetic analyses of Synechocystis sp. PCC6803 and Chlorogloeopsis fritschii sp. PCC6912 exposed to an M-dwarf spectrum under an anoxic atmosphere
Abstract
Introduction: Cyanobacteria appeared in the anoxic Archean Earth, evolving for the first time oxygenic photosynthesis and deeply changing the atmosphere by introducing oxygen. Starting possibly from UV-protected environments, characterized by low visible and far-red enriched light spectra, cyanobacteria spread everywhere on Earth thanks to their adaptation capabilities in light harvesting. In the last decade, few cyanobacteria species which can acclimate to far-red light through Far-Red Light Photoacclimation (FaRLiP) have been isolated. FaRLiP cyanobacteria were thus proposed as model organisms to study the origin of oxygenic photosynthesis as well as its possible functionality around stars with high far-red emission, the M-dwarfs. These stars are astrobiological targets, as their longevity could sustain life evolution and they demonstrated to host rocky terrestrial-like exoplanets within their Habitable Zone.
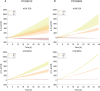
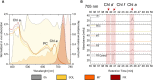
Methods: We studied the acclimation responses of the FaRLiP strain Chlorogloeopsis fritschii sp. PCC6912 and the non-FaRLiP strain Synechocystis sp. PCC6803 to the combination of three simulated light spectra (M-dwarf, solar and far-red) and two atmospheric compositions (oxic, anoxic). We first checked their growth, O2 production and pigment composition, then we studied their transcriptional responses by RNA sequencing under each combination of light spectrum and atmosphere conditions.
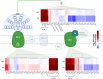
Results and discussion: PCC6803 did not show relevant differences in gene expression when comparing the responses to M-dwarf and solar-simulated lights, while far-red caused a variation in the transcriptional level of many genes. PCC6912 showed, on the contrary, different transcriptional responses to each light condition and activated the FaRLiP response under the M-dwarf simulated light. Surprisingly, the anoxic atmosphere did not impact the transcriptional profile of the 2 strains significantly. Results show that both cyanobacteria seem inherently prepared for anoxia and to harvest the photons emitted by a simulated M-dwarf star, whether they are only visible (PCC6803) or also far-red photons (PCC6912). They also show that visible photons in the simulated M-dwarf are sufficient to keep a similar metabolism with respect to solar-simulated light.
Conclusion: Results prove the adaptability of the cyanobacterial metabolism and enhance the plausibility of finding oxygenic biospheres on exoplanets orbiting M-dwarf stars.
Keywords: FaRLiP; M-dwarf spectrum; RNA-Seq; anoxia; cyanobacteria; laboratory simulation experiments; transcriptomic analysis.
Copyright © 2024 Battistuzzi, Morlino, Cocola, Trainotti, Treu, Campanaro, Claudi, Poletto and La Rocca.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


References
-
- Adams F. C., Laughlin G., Graves G. J. M. (2004). Red dwarfs and the end of the main sequence. Rev. Mexicana Astronomia Y Astrofisica: Serie Conferencias 22, 46–49. Available at: https://www.redalyc.org/articulo.oa?id=57102211.
-
- Airs R. L., Temperton B., Sambles C., Farnham G., Skill S. C., Llewellyn C. A. (2014). Chlorophyll f and chlorophyll d are produced in the cyanobacterium Chlorogloeopsis fritschii when cultured under natural light and near-infrared radiation. FEBS Lett. 588 (20), 3770–3777. doi: 10.1016/j.febslet.2014.08.026. Federation of European Biochemical Societies. - DOI - PubMed
-
- Battistuzzi M., Cocola L., Salasnich B., Erculiani M. S., Alei E., Morosinotto T., et al. . (2020). A new remote sensing-based system for the monitoring and analysis of growth and gas exchange rates of photosynthetic microorganisms under simulated non-terrestrial conditions. Front. Plant Sci. 11, 182. doi: 10.3389/fpls.2020.00182 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

