Disenfranchised DNA: biochemical analysis of mutant øX174 DNA-binding proteins may further elucidate the evolutionary significance of the unessential packaging protein A
- PMID: 38305183
- PMCID: PMC10949513
- DOI: 10.1128/jvi.01827-23
Disenfranchised DNA: biochemical analysis of mutant øX174 DNA-binding proteins may further elucidate the evolutionary significance of the unessential packaging protein A
Abstract
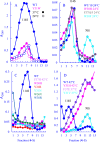
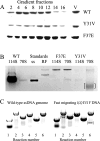
Most icosahedral DNA viruses package and condense their genomes into pre-formed, volumetrically constrained capsids. However, concurrent genome biosynthesis and packaging are specific to single-stranded (ss) DNA micro- and parvoviruses. Before packaging, ~120 copies of the øX174 DNA-binding protein J interact with double-stranded DNA. 60 J proteins enter the procapsid with the ssDNA genome, guiding it between 60 icosahedrally ordered DNA-binding pockets formed by the capsid proteins. Although J proteins are small, 28-37 residues in length, they have two domains. The basic, positively charged N-terminus guides the genome between binding pockets, whereas the C-terminus acts as an anchor to the capsid's inner surface. Three C-terminal aromatic residues, W30, Y31, and F37, interact most extensively with the coat protein. Their corresponding codons were mutated, and the resulting strains were biochemically and genetically characterized. Depending on the mutation, the substitutions produced unstable packaging complexes, unstable virions, infectious progeny, or particles packaged with smaller genomes, the latter being a novel phenomenon. The smaller genomes contained internal deletions. The juncture sequences suggest that the unessential A* (A star) protein mediates deletion formation.IMPORTANCEUnessential but strongly conserved gene products are understudied, especially when mutations do not confer discernable phenotypes or the protein's contribution to fitness is too small to reliably determine in laboratory-based assays. Consequently, their functions and evolutionary impact remain obscure. The data presented herein suggest that microvirus A* proteins, discovered over 40 years ago, may hasten the termination of non-productive packaging events. Thus, performing a salvage function by liberating the reusable components of the failed packaging complexes, such as DNA templates and replication enzymes.
Keywords: microviridae; phiX174; single-stranded DNA packaging; single-stranded DNA replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
The Effects of Packaged, but Misguided, Single-Stranded DNA Genomes Are Transmitted to the Outer Surface of the ϕX174 Capsid.J Virol. 2021 Aug 25;95(18):e0088321. doi: 10.1128/JVI.00883-21. Epub 2021 Aug 25. J Virol. 2021. PMID: 34232738 Free PMC article.
-
Finally, a Role Befitting Astar: Strongly Conserved, Unessential Microvirus A* Proteins Ensure the Product Fidelity of Packaging Reactions.J Virol. 2020 Jan 6;94(2):e01593-19. doi: 10.1128/JVI.01593-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31666371 Free PMC article.
-
Snow goose hepatitis B virus (SGHBV) envelope and capsid proteins independently contribute to the ability of SGHBV to package capsids containing single-stranded DNA in virions.J Virol. 2014 Sep;88(18):10705-13. doi: 10.1128/JVI.01694-14. Epub 2014 Jul 2. J Virol. 2014. PMID: 24991016 Free PMC article.
-
A Molecular Staple: D-Loops in the I Domain of Bacteriophage P22 Coat Protein Make Important Intercapsomer Contacts Required for Procapsid Assembly.J Virol. 2015 Oct;89(20):10569-79. doi: 10.1128/JVI.01629-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269173 Free PMC article.
-
Mysteries of adenovirus packaging.J Virol. 2025 May 20;99(5):e0018025. doi: 10.1128/jvi.00180-25. Epub 2025 Apr 17. J Virol. 2025. PMID: 40243339 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

