Multimodal immune phenotyping reveals microbial-T cell interactions that shape pancreatic cancer
- PMID: 38307029
- PMCID: PMC10897543
- DOI: 10.1016/j.xcrm.2024.101397
Multimodal immune phenotyping reveals microbial-T cell interactions that shape pancreatic cancer
Abstract
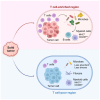
Microbes are an integral component of the tumor microenvironment. However, determinants of microbial presence remain ill-defined. Here, using spatial-profiling technologies, we show that bacterial and immune cell heterogeneity are spatially coupled. Mouse models of pancreatic cancer recapitulate the immune-microbial spatial coupling seen in humans. Distinct intra-tumoral niches are defined by T cells, with T cell-enriched and T cell-poor regions displaying unique bacterial communities that are associated with immunologically active and quiescent phenotypes, respectively, but are independent of the gut microbiome. Depletion of intra-tumoral bacteria slows tumor growth in T cell-poor tumors and alters the phenotype and presence of myeloid and B cells in T cell-enriched tumors but does not affect T cell infiltration. In contrast, T cell depletion disrupts the immunological state of tumors and reduces intra-tumoral bacteria. Our results establish a coupling between microbes and T cells in cancer wherein spatially defined immune-microbial communities differentially influence tumor biology.
Keywords: T cells; bacteria; cellular communities; gut microbiome; immune system; lung adenocarcinoma; pancreatic ductal adenocarcinoma; spatial heterogeneity; tumor microbiome; tumor microenvironment.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests W.J.H. reports royalties from Rodeo/Amgen; grants from Sanofi, NeoTX, and Circle Pharma; and prior consulting fees from Exelixis. G.L.B. reports prior and active roles as a consultant/advisory board member for Seattle Genetics (now Seagen), Adicet Bio, Aduro Biotech, AstraZeneca, BiolineRx, BioMarin Pharmaceuticals, Bristol-Myers Squibb, Cantargia, Cour Pharmaceuticals, Boehinger Ingelheim, Genmab, Hibercell, HotSpot Therapeutics, Incyte Corporation, Janssen, Merck, Molecular Partners, NanoGhost, Pancreatic Cancer Action Network, Shattuck Labs, and Verastem and reports receiving commercial research grants from Incyte Corporation, Bristol-Myers Squibb, Verastem, Halozyme, Biothera, Newlink, Novartis, Arcus Biosciences, and Janssen. G.L.B. is an inventor of intellectual property related to chimeric antigen receptor (CAR) T cells that is licensed by the University of Pennsylvania to Novartis and Tmunity Therapeutics.
Figures





References
-
- Fu A., Yao B., Dong T., Chen Y., Yao J., Liu Y., Li H., Bai H., Liu X., Zhang Y., et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. 2022;185:1356–1372.e26. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

