Human ACVR1C missense variants that correlate with altered body fat distribution produce metabolic alterations of graded severity in knock-in mutant mice
- PMID: 38307384
- PMCID: PMC10863331
- DOI: 10.1016/j.molmet.2024.101890
Human ACVR1C missense variants that correlate with altered body fat distribution produce metabolic alterations of graded severity in knock-in mutant mice
Abstract
Background & aims: Genome-wide studies have identified three missense variants in the human gene ACVR1C, encoding the TGF-β superfamily receptor ALK7, that correlate with altered waist-to-hip ratio adjusted for body mass index (WHR/BMI), a measure of body fat distribution.
Methods: To move from correlation to causation and understand the effects of these variants on fat accumulation and adipose tissue function, we introduced each of the variants in the mouse Acvr1c locus and investigated metabolic phenotypes in comparison with a null mutation.
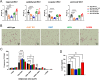
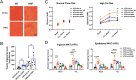
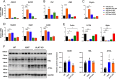
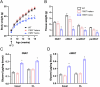
Results: Mice carrying the I195T variant showed resistance to high fat diet (HFD)-induced obesity, increased catecholamine-induced adipose tissue lipolysis and impaired ALK7 signaling, phenocopying the null mutants. Mice with the I482V variant displayed an intermediate phenotype, with partial resistance to HFD-induced obesity, reduction in subcutaneous, but not visceral, fat mass, decreased systemic lipolysis and reduced ALK7 signaling. Surprisingly, mice carrying the N150H variant were metabolically indistinguishable from wild type under HFD, although ALK7 signaling was reduced at low ligand concentrations.
Conclusion: Together, these results validate ALK7 as an attractive drug target in human obesity and suggest a lower threshold for ALK7 function in humans compared to mice.
Keywords: Adipose tissue; Lipolysis; Obesity; Single-nucleotide variant.
Copyright © 2024 The Author(s). Published by Elsevier GmbH.. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have influenced the results reported in this paper.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

