Lung endothelium exploits susceptible tumor cell states to instruct metastatic latency
- PMID: 38308117
- PMCID: PMC11136671
- DOI: 10.1038/s43018-023-00716-7
Lung endothelium exploits susceptible tumor cell states to instruct metastatic latency
Abstract
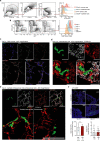
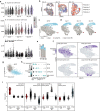
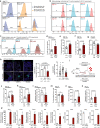
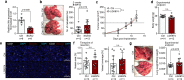
In metastasis, cancer cells travel around the circulation to colonize distant sites. Due to the rarity of these events, the immediate fates of metastasizing tumor cells (mTCs) are poorly understood while the role of the endothelium as a dissemination interface remains elusive. Using a newly developed combinatorial mTC enrichment approach, we provide a transcriptional blueprint of the early colonization process. Following their arrest at the metastatic site, mTCs were found to either proliferate intravascularly or extravasate, thereby establishing metastatic latency. Endothelial-derived angiocrine Wnt factors drive this bifurcation, instructing mTCs to follow the extravasation-latency route. Surprisingly, mTC responsiveness towards niche-derived Wnt was established at the epigenetic level, which predetermined tumor cell behavior. Whereas hypomethylation enabled high Wnt activity leading to metastatic latency, methylated mTCs exhibited low activity and proliferated intravascularly. Collectively the data identify the predetermined methylation status of disseminated tumor cells as a key regulator of mTC behavior in the metastatic niche.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- 39404578/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 331351713/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 787181/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- Angiomature/Deutsche Krebshilfe (German Cancer Aid)
- 00.022.2019/Klaus Tschira Stiftung (Klaus Tschira Foundation)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

