NAMPT-Driven M2 Polarization of Tumor-Associated Macrophages Leads to an Immunosuppressive Microenvironment in Colorectal Cancer
- PMID: 38308188
- PMCID: PMC11005718
- DOI: 10.1002/advs.202303177
NAMPT-Driven M2 Polarization of Tumor-Associated Macrophages Leads to an Immunosuppressive Microenvironment in Colorectal Cancer
Abstract
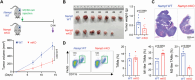
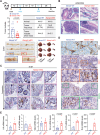
Nicotinamide phosphoribosyltransferase (NAMPT) is a metabolic enzyme with key roles in inflammation. Previous studies have examined the consequences of its upregulated expression in cancer cells themselves, but studies are limited with respect to its role in the other cells within the tumor microenvironment (TME) during colorectal cancer (CRC) progression. Using single-cell RNA sequencing (scRNA-seq) data, it is founded that NAMPT is highly expressed in SPP1+ tumor-associated macrophages (TAMs), a unique subset of TAMs associated with immunosuppressive activity. A NAMPThigh gene signature in SPP1+ TAMs correlated with worse prognostic outcomes in CRC patients. The effect of Nampt deletion in the myeloid compartment of mice during CRC development is explored. NAMPT deficiency in macrophages resulted in HIF-1α destabilization, leading to reduction in M2-like TAM polarization. NAMPT deficiency caused significant decreases in the efferocytosis activity of macrophages, which enhanced STING signaling and the induction of type I IFN-response genes. Expression of these genes contributed to anti-tumoral immunity via potentiation of cytotoxic T cell activity in the TME. Overall, these findings suggest that NAMPT-initiated TAM-specific genes can be useful in predicting poor CRC patient outcomes; strategies aimed at targeting NAMPT may provide a promising therapeutic approach for building an immunostimulatory TME in CRC progression.
Keywords: CRC; HIF‐1α; M2‐like TAMs; NAMPT; STING.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
