Zygotic Splicing Activation of the Transcriptome is a Crucial Aspect of Maternal-to-Zygotic Transition and Required for the Conversion from Totipotency to Pluripotency
- PMID: 38308190
- PMCID: PMC11005748
- DOI: 10.1002/advs.202308496
Zygotic Splicing Activation of the Transcriptome is a Crucial Aspect of Maternal-to-Zygotic Transition and Required for the Conversion from Totipotency to Pluripotency
Abstract
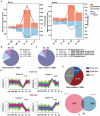
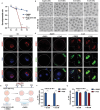
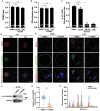
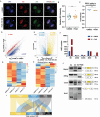
During maternal-to-zygotic transition (MZT) in the embryo, mRNA undergoes complex post-transcriptional regulatory processes. However, it is unclear whether and how alternative splicing plays a functional role in MZT. By analyzing transcriptome changes in mouse and human early embryos, dynamic changes in alternative splicing during MZT are observed and a previously unnoticed process of zygotic splicing activation (ZSA) following embryonic transcriptional activation is described. As the underlying mechanism of RNA splicing, splicing factors undergo dramatic maternal-to-zygotic conversion. This conversion relies on the key maternal factors BTG4 and PABPN1L and is zygotic-transcription-dependent. CDK11-dependent phosphorylation of the key splicing factor, SF3B1, and its aggregation with SRSF2 in the subnuclear domains of 2-cell embryos are prerequisites for ZSA. Isoforms generated by erroneous splicing, such as full-length Dppa4, hinder normal embryonic development. Moreover, alternative splicing regulates the conversion of early embryonic blastomeres from totipotency to pluripotency, thereby affecting embryonic lineage differentiation. ZSA is an essential post-transcriptional process of MZT and has physiological significance in generating new life. In addition to transcriptional activation, appropriate expression of transcript isoforms is also necessary for preimplantation embryonic development.
Keywords: RNA processing; alternative splicing; early embryo; splicing factors; totipotency and pluripotency; zygotic genome activation.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
- 2021YFC2700100/National Key Research and Development Program of China
- 2021C03100/Key Research and Development Program of Zhejiang Province
- 2021C03098/Key Research and Development Program of Zhejiang Province
- 31930031/National Natural Science Foundation of China
- 32072939/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Research Materials
