PHLDA2-mediated phosphatidic acid peroxidation triggers a distinct ferroptotic response during tumor suppression
- PMID: 38309267
- PMCID: PMC11209835
- DOI: 10.1016/j.cmet.2024.01.006
PHLDA2-mediated phosphatidic acid peroxidation triggers a distinct ferroptotic response during tumor suppression
Abstract
Although the role of ferroptosis in killing tumor cells is well established, recent studies indicate that ferroptosis inducers also sabotage anti-tumor immunity by killing neutrophils and thus unexpectedly stimulate tumor growth, raising a serious issue about whether ferroptosis effectively suppresses tumor development in vivo. Through genome-wide CRISPR-Cas9 screenings, we discover a pleckstrin homology-like domain family A member 2 (PHLDA2)-mediated ferroptosis pathway that is neither ACSL4-dependent nor requires common ferroptosis inducers. PHLDA2-mediated ferroptosis acts through the peroxidation of phosphatidic acid (PA) upon high levels of reactive oxygen species (ROS). ROS-induced ferroptosis is critical for tumor growth in the absence of common ferroptosis inducers; strikingly, loss of PHLDA2 abrogates ROS-induced ferroptosis and promotes tumor growth but has no obvious effect in normal tissues in both immunodeficient and immunocompetent mouse tumor models. These data demonstrate that PHLDA2-mediated PA peroxidation triggers a distinct ferroptosis response critical for tumor suppression and reveal that PHLDA2-mediated ferroptosis occurs naturally in vivo without any treatment from ferroptosis inducers.
Keywords: ALOX12; GPAT3; PHLDA2; ROS; cystine starvation; ferroptosis; lipid peroxidation; phosphatidic acid; phosphatidylethanolamine; tumor suppression.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.R.S. is an inventor on patents and patent applications involving small molecule drug discovery and ferroptosis; holds equity in Sonata Therapeutics; holds equity in and serves as a consultant to Exarta Therapeutics and ProJenX, Inc.; and serves as a consultant to Weatherwax Biotechnologies Corporation and Akin Gump Strauss Hauer & Feld LLP. X.J. is an inventor on patents related to autophagy and cell death and holds equity of and consults for Exarta Therapeutics and Lime Therapeutics.
Figures

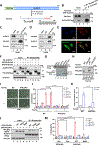
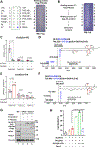
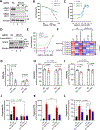

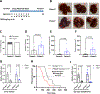
References
-
- Chen X, Kang R, Kroemer G, and Tang D Broadening horizons: the role of ferroptosis in cancer. Nature reviews Clinical oncology 18, 280–296 (2021). - PubMed
MeSH terms
Substances
Grants and funding
- R01 CA165065/CA/NCI NIH HHS/United States
- U01 AI156924/AI/NIAID NIH HHS/United States
- R01 NS076511/NS/NINDS NIH HHS/United States
- R01 NS061817/NS/NINDS NIH HHS/United States
- P30 CA013696/CA/NCI NIH HHS/United States
- R01 CA258622/CA/NCI NIH HHS/United States
- R35 CA253059/CA/NCI NIH HHS/United States
- R01 CA204232/CA/NCI NIH HHS/United States
- R01 CA258390/CA/NCI NIH HHS/United States
- R01 CA254970/CA/NCI NIH HHS/United States
- R35 CA209896/CA/NCI NIH HHS/United States
- R37 NS061817/NS/NINDS NIH HHS/United States
- R01 CA266342/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- U01 AI156923/AI/NIAID NIH HHS/United States
- R01 CA243142/CA/NCI NIH HHS/United States
- R01 CA166413/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

