Phosphoglycerate kinase 1 acts as a cargo adaptor to promote EGFR transport to the lysosome
- PMID: 38310114
- PMCID: PMC10838266
- DOI: 10.1038/s41467-024-45443-4
Phosphoglycerate kinase 1 acts as a cargo adaptor to promote EGFR transport to the lysosome
Abstract
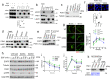
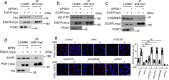
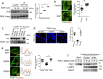
The epidermal growth factor receptor (EGFR) plays important roles in multiple cellular events, including growth, differentiation, and motility. A major mechanism of downregulating EGFR function involves its endocytic transport to the lysosome. Sorting of proteins into intracellular pathways involves cargo adaptors recognizing sorting signals on cargo proteins. A dileucine-based sorting signal has been identified previously for the sorting of endosomal EGFR to the lysosome, but a cargo adaptor that recognizes this signal remains unknown. Here, we find that phosphoglycerate kinase 1 (PGK1) is recruited to endosomal membrane upon its phosphorylation, where it binds to the dileucine sorting signal in EGFR to promote the lysosomal transport of this receptor. We also elucidate two mechanisms that act in concert to promote PGK1 recruitment to endosomal membrane, a lipid-based mechanism that involves phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] and a protein-based mechanism that involves hepatocyte growth factor receptor substrate (Hrs). These findings reveal an unexpected function for a metabolic enzyme and advance the mechanistic understanding of how EGFR is transported to the lysosome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

