Impact of KRAS mutations and co-mutations on clinical outcomes in pancreatic ductal adenocarcinoma
- PMID: 38310130
- PMCID: PMC10838312
- DOI: 10.1038/s41698-024-00505-0
Impact of KRAS mutations and co-mutations on clinical outcomes in pancreatic ductal adenocarcinoma
Abstract
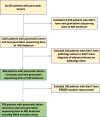
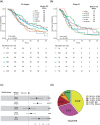
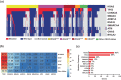
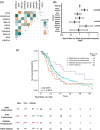
The relevance of KRAS mutation alleles to clinical outcome remains inconclusive in pancreatic adenocarcinoma (PDAC). We conducted a retrospective study of 803 patients with PDAC (42% with metastatic disease) at MD Anderson Cancer Center. Overall survival (OS) analysis demonstrated that KRAS mutation status and subtypes were prognostic (p < 0.001). Relative to patients with KRAS wildtype tumors (median OS 38 months), patients with KRASG12R had a similar OS (median 34 months), while patients with KRASQ61 and KRASG12D mutated tumors had shorter OS (median 20 months [HR: 1.9, 95% CI 1.2-3.0, p = 0.006] and 22 months [HR: 1.7, 95% CI 1.3-2.3, p < 0.001], respectively). There was enrichment of KRASG12D mutation in metastatic tumors (34% vs 24%, OR: 1.7, 95% CI 1.2-2.4, p = 0.001) and enrichment of KRASG12R in well and moderately differentiated tumors (14% vs 9%, OR: 1.7, 95% CI 1.05-2.99, p = 0.04). Similar findings were observed in the external validation cohort (PanCAN's Know Your Tumor® dataset, n = 408).
© 2024. The Author(s).
Conflict of interest statement
D.Z. has clinical trial contracts with Mirati and CARsgen and served as a member of the advisory board for Affini-T.K.A., S.D., and L.M. are employees of the Pancreatic Cancer Action Network, a nonprofit cancer advocacy organization that receives donations from private and commercial entities and may benefit indirectly financially and non-financially from this publication. S.P. reports research funding from Mirati Therapeutics, Lilly, Xencor, Novartis, Rgenix, Bristol-Myers Squibb, Astellas Pharma, Framewave, 4D Pharma, Boehringer Ingelheim, NGM Biopharmaceuticals, Janssen, Arcus Biosciences, Elicio Therapeutics, Bionote, Ipsen, Zymeworks, Pfizer, ImmunoMET, Immuneering, Amal Therapeutics. S.P. is a member of the advisory board for Zymeworks, Ipsen, Novartis, Janssen, Boehringer Ingelheim, AskGene Pharma, BPGbio, Jazz Pharmaceuticals, AstraZeneca, US WorldMeds, Nihon Medi-Physics Co., Ltd., Alligator Bioscience. M.L. consults for Pfizer, Delcath, Janssen, BioNTech, G1 Therapeutics, Imvax, and Bayer and received research funding from institutions from EpimAb BioTherapeutics, Merck, Erasca, Boehringer Ingelheim, Arcus Biosciences, Repare Therapeutics, Trisalus Life Sciences, and Xilis. A.M. is a consultant for Tezcat Biotechnologies and is a co-inventor on a patent that has been licensed from Johns Hopkins University by Thrive Earlier Detection (an Exact Sciences company). A.M. serves on the scientific and medical advisory board for Pancreatic Cancer Action Network and Pancreatic Cancer UK. J.P.S. receives grants, research support, or collaborates with Celsius Therapeutics, BostonGene, Caris Life Sciences, Natera, Xilis, Palantir, and Genentech. J.P.S. reports consulting and stock ownership with Engine Biosciences, NaDeNo Nanoscience.
Figures


Update of
-
Impact of KRAS Mutations and Co-mutations on Clinical Outcomes in Pancreatic Ductal Adenocarcinoma.Res Sq [Preprint]. 2023 Aug 10:rs.3.rs-3195257. doi: 10.21203/rs.3.rs-3195257/v1. Res Sq. 2023. Update in: NPJ Precis Oncol. 2024 Feb 3;8(1):27. doi: 10.1038/s41698-024-00505-0. PMID: 37609177 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

