Epithelial overexpression of IL-33 induces eosinophilic esophagitis dependent on IL-13
- PMID: 38310974
- PMCID: PMC11070306
- DOI: 10.1016/j.jaci.2024.01.017
Epithelial overexpression of IL-33 induces eosinophilic esophagitis dependent on IL-13
Abstract
Background: Eosinophilic esophagitis (EoE) is an increasingly common inflammatory condition of the esophagus; however, the underlying immunologic mechanisms remain poorly understood. The epithelium-derived cytokine IL-33 is associated with type 2 immune responses and elevated in esophageal biopsy specimens from patients with EoE.
Objective: We hypothesized that overexpression of IL-33 by the esophageal epithelium would promote the immunopathology of EoE.
Methods: We evaluated the functional consequences of esophageal epithelial overexpression of a secreted and active form of IL-33 in a novel transgenic mouse, EoE33. EoE33 mice were analyzed for clinical and immunologic phenotypes. Esophageal contractility was assessed. Epithelial cytokine responses were analyzed in three-dimensional organoids. EoE33 phenotypes were further characterized in ST2-/-, eosinophil-deficient, and IL-13-/- mice. Finally, EoE33 mice were treated with dexamethasone.
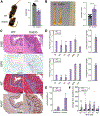
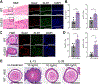
Results: EoE33 mice displayed ST2-dependent, EoE-like pathology and failed to thrive. Esophageal tissue remodeling and inflammation included basal zone hyperplasia, eosinophilia, mast cells, and TH2 cells. Marked increases in levels of type 2 cytokines, including IL-13, and molecules associated with immune responses and tissue remodeling were observed. Esophageal organoids suggested reactive epithelial changes. Genetic deletion of IL-13 in EoE33 mice abrogated pathologic changes in vivo. EoE33 mice were responsive to steroids.
Conclusions: IL-33 overexpression by the esophageal epithelium generated immunopathology and clinical phenotypes resembling human EoE. IL-33 may play a pivotal role in the etiology of EoE by activating the IL-13 pathway. EoE33 mice are a robust experimental platform for mechanistic investigation and translational discovery.
Keywords: IL-33; eosinophil; eosinophilic esophagitis; transgene; type 2 inflammation.
Copyright © 2024 American Academy of Allergy, Asthma & Immunology. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Simon D, Radonjic-Hosli S, Straumann A, Yousefi S, Simon HU. Active eosinophilic esophagitis is characterized by epithelial barrier defects and eosinophil extracellular trap formation. Allergy 2015; 70:443–52. - PubMed
-
- Judd LM, Heine RG, Menheniott TR, Buzzelli J, O’Brien-Simpson N, Pavlic D, et al. Elevated IL-33 expression is associated with pediatric eosinophilic esophagitis, and exogenous IL-33 promotes eosinophilic esophagitis development in mice. Am J Physiol Gastrointest Liver Physiol 2016; 310:G13–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

