DLK1/DIO3 locus upregulation by a β-catenin-dependent enhancer drives cell proliferation and liver tumorigenesis
- PMID: 38311851
- PMCID: PMC11163201
- DOI: 10.1016/j.ymthe.2024.01.036
DLK1/DIO3 locus upregulation by a β-catenin-dependent enhancer drives cell proliferation and liver tumorigenesis
Abstract
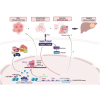
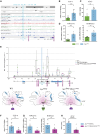
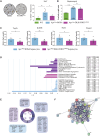
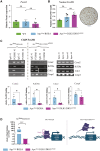
The CTNNB1 gene, encoding β-catenin, is frequently mutated in hepatocellular carcinoma (HCC, ∼30%) and in hepatoblastoma (HB, >80%), in which DLK1/DIO3 locus induction is correlated with CTNNB1 mutations. Here, we aim to decipher how sustained β-catenin activation regulates DLK1/DIO3 locus expression and the role this locus plays in HB and HCC development in mouse models deleted for Apc (ApcΔhep) or Ctnnb1-exon 3 (β-cateninΔExon3) and in human CTNNB1-mutated hepatic cancer cells. We identified an enhancer site bound by TCF-4/β-catenin complexes in an open conformation upon sustained β-catenin activation (DLK1-Wnt responsive element [WRE]) and increasing DLK1/DIO3 locus transcription in β-catenin-mutated human HB and mouse models. DLK1-WRE editing by CRISPR-Cas9 approach impaired DLK1/DIO3 locus expression and slowed tumor growth in subcutaneous CTNNB1-mutated tumor cell grafts, ApcΔhep HB and β-cateninΔExon3 HCC. Tumor growth inhibition resulted either from increased FADD expression and subsequent caspase-3 cleavage in the first case or from decreased expression of cell cycle actors regulated by FoxM1 in the others. Therefore, the DLK1/DIO3 locus is an essential determinant of FoxM1-dependent cell proliferation during β-catenin-driven liver tumorigenesis. Targeting the DLK1-WRE enhancer to silence the DLK1/DIO3 locus might thus represent an interesting therapeutic strategy to restrict tumor growth in primary liver cancers with CTNNB1 mutations.
Keywords: enhancer site; in vivo CRISPR-Cas9; non-coding RNAs; primary liver cancers; targeted therapies; transgenic mice; β-catenin.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Two patents, PCT/EP2023/053419 and EP22305162.4, have been deposited by J.S., L.P., S.C., and A.G.
Figures




References
-
- Hirsch T.Z., Pilet J., Morcrette G., Roehrig A., Monteiro B.J.E., Molina L., Bayard Q., Trépo E., Meunier L., Caruso S., et al. Integrated Genomic Analysis Identifies Driver Genes and Cisplatin-Resistant Progenitor Phenotype in Pediatric Liver Cancer. Cancer Discov. 2021;11:2524–2543. doi: 10.1158/2159-8290.CD-20-1809. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

