Allosteric Signal within the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein Mediated by a Class 3 Monoclonal Antibody Revealed through Molecular Dynamics Simulations and Protein Residue Networks
- PMID: 38313482
- PMCID: PMC10831861
- DOI: 10.1021/acsomega.3c07947
Allosteric Signal within the Receptor-Binding Domain of the SARS-CoV-2 Spike Protein Mediated by a Class 3 Monoclonal Antibody Revealed through Molecular Dynamics Simulations and Protein Residue Networks
Abstract
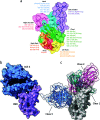
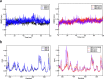
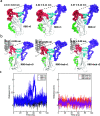
This study investigated the allosteric action within the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein caused by class 3 monoclonal antibody (mAb) binding. As the emergence of SARS-CoV-2 variants has raised concerns about the effectiveness of treatments by antibodies, targeting the highly conserved class 3 epitopes has become an alternative strategy of antibody design. Simulations of explicitly solvated RBD of the BA.2.75 omicron subvariants were carried out both in the presence and in the absence of bebtelovimab, as a model example of class 3 monoclonal antibodies against the RBD of the SARS-CoV-2 spike protein. The comparative analysis showed that bebtelovimab's binding on two α helices at the epitope region disrupted the nearby interaction network, which triggered a denser interaction network formation on the opposite side of the receptor-binding motif (RBM) region and resulted in a "close" conformation that could prevent the ACE2 binding. A better understanding of this allosteric action could lead to the development of alternative mAbs for further variants of concern. In terms of computational techniques, the communicability matrix could serve as a tool to visualize the effects of allostery, as the pairs of amino acids or secondary structures with high communicability could pinpoint the possible sites to transfer the allosteric signal. Additionally, the communicability gain/loss matrix could help elucidate the consequences of allosteric actions, which could be employed along with other allostery quantification techniques in some previous studies.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
