Association of HBsAg levels with differential gene expression in NK, CD8 T, and memory B cells in treated patients with chronic HBV
- PMID: 38314025
- PMCID: PMC10835465
- DOI: 10.1016/j.jhepr.2023.100980
Association of HBsAg levels with differential gene expression in NK, CD8 T, and memory B cells in treated patients with chronic HBV
Abstract
Background & aims: HBsAg secretion may impact immune responses to chronic HBV infection. Thus, therapeutic approaches to suppress HBsAg production are being investigated. Our study aims to examine the immunomodulatory effects of high and low levels of circulating HBsAg and thereby improve our understanding of anti-HBV immunity.
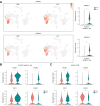
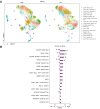
Methods: An optimized 10x Genomics single-cell RNA sequencing workflow was applied to blood samples and liver fine-needle aspirates from 18 patients undergoing tenofovir/entecavir (NUC) treatment for chronic HBV infection. They were categorized based on their HBsAg levels: high (920-12,447 IU/ml) or low (1-100 IU/ml). Cluster frequencies, differential gene expression, and phenotypes were analyzed.
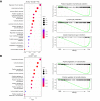
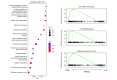
Results: In the blood of HBV-infected patients on NUC, the proportion of KLRC2+ "adaptive" natural killer (NK) cells was significantly lower in the HBsAg-high group and, remarkably, both KLRC2+ NK and KLRG1+ CD8 T cells display enrichment of lymphocyte activation-associated gene sets in the HBsAg-low group. High levels of HBsAg were associated with mild immune activation in the liver. However, no suppression of liver-resident CXCR6+ NCAM1+ NK or CXCR6+ CD69+ CD8 T cells was detected, while memory B cells showed signs of activation in both the blood and liver.
Conclusions: Among NUC-treated patients, we observed a minimal impact of HBsAg on leukocyte populations in the blood and liver. However, for the first time, we found that HBsAg has distinct effects, restricted to NK-, CD8 T-, and memory B-cell subsets, in the blood and liver. Our findings are highly relevant for current clinical studies evaluating treatment strategies aimed at suppressing HBsAg production and reinvigorating immunity to HBV.
Impact and implications: This study provides unique insight into the impact of HBsAg on gene expression levels of immune cell subsets in the blood and liver, particularly in the context of NUC-treated chronic HBV infection. It holds significant relevance for current and future clinical studies evaluating treatment strategies aimed at suppressing HBsAg production and reinvigorating immunity to HBV. Our findings raise questions about the effectiveness of such treatment strategies and challenge the previously hypothesized immunomodulatory effects of HBsAg on immune responses against HBV.
Keywords: HBsAg; hepatitis B; liver fine-needle aspirates; single-cell RNA sequencing.
© 2023 The Author(s).
Conflict of interest statement
A.B. received grants not related to this project from Gilead Sciences, Fujirebio, GlaxoSmithKline, and Janssen Pharma. R.J.K. received grants from GlaxoSmithKline, Janssen-Cilag, and Echosens (not related to this project), and received consulting fees, payments or honoraria for lectures, presentations, or other educational events from AbbVie, Echosens, and Gilead Sciences. All remaining authors declare that they have no conflicts of interest. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures




References
-
- McLane L.M., Abdel-Hakeem M.S., Wherry E.J. CD8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol. 2019;37:457–495. - PubMed
-
- Tout I., Loureiro D., Mansouri A., et al. Hepatitis B surface antigen seroclearance: immune mechanisms, clinical impact, importance for drug development. J Hepatol. 2020;73:409–422. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

