Engineering lentivirus envelope VSV-G for liver targeted delivery of IDOL-shRNA to ameliorate hypercholesterolemia and atherosclerosis
- PMID: 38314097
- PMCID: PMC10835450
- DOI: 10.1016/j.omtn.2024.102115
Engineering lentivirus envelope VSV-G for liver targeted delivery of IDOL-shRNA to ameliorate hypercholesterolemia and atherosclerosis
Abstract
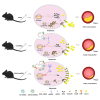
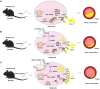
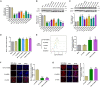
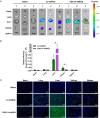
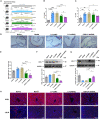
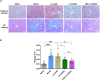
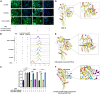
Lentiviral vectors (LVs) have been widely used as a tool for gene therapies. However, tissue-selective transduction after systemic delivery remains a challenge. Inducible degrader of low-density lipoprotein receptor is an attractive target for treating hypercholesterolemia. Here, a liver-targeted LV, CS8-LV-shIDOL, is developed by incorporating a hepatocyte-targeted peptide derived from circumsporozoite protein (CSP) into the lentivirus envelope for liver-targeted delivery of IDOL-shRNA (short hairpin RNA) to alleviate hypercholesterolemia. Tail-vein injection of CS8-LV-shIDOL results in extremely high accumulation in liver and nearly undetectable levels in other organs in mice. In addition, it shows superior therapeutic efficacy in lowering serum low-density lipoprotein cholesterol (LDL-C) and reducing atherosclerotic lesions over unmodified LV-shIDOL in hyperlipidemic mice. Mechanically, the envelope-engineered CS8-LV-shIDOL can enter liver cells via low-density lipoprotein receptor-related protein (LRP). Thus, this study provides a novel approach for liver-targeted delivery of IDOL-shRNA to treat hypercholesterolemia by using an envelope-engineered LV, and this delivery system has great potential for liver-targeted transgene therapy.
Keywords: IDOL; MT: Oligonucleotides: Therapies and Applications; gene therapy; lentivirus; liver-targeted; siRNA; small interfering RNA.
© 2024 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Stable liver-specific expression of human IDOL in humanized mice raises plasma cholesterol.Cardiovasc Res. 2016 May 1;110(1):23-9. doi: 10.1093/cvr/cvw010. Epub 2016 Jan 19. Cardiovasc Res. 2016. PMID: 26786161 Free PMC article.
-
Idol Depletion Protects against Spontaneous Atherosclerosis in a Hamster Model of Familial Hypercholesterolemia.Oxid Med Cell Longev. 2022 May 24;2022:1889632. doi: 10.1155/2022/1889632. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35656026 Free PMC article.
-
Local lentiviral short hairpin RNA silencing of CCR2 inhibits vein graft thickening in hypercholesterolemic apolipoprotein E3-Leiden mice.J Vasc Surg. 2009 Jul;50(1):152-60. doi: 10.1016/j.jvs.2009.03.027. J Vasc Surg. 2009. PMID: 19563963
-
Post-transcriptional regulation of lipoprotein receptors by the E3-ubiquitin ligase inducible degrader of the low-density lipoprotein receptor.Curr Opin Lipidol. 2012 Jun;23(3):213-219. doi: 10.1097/MOL.0b013e3283532947. Curr Opin Lipidol. 2012. PMID: 22510808 Review.
-
Feedback regulation of cholesterol uptake by the LXR-IDOL-LDLR axis.Arterioscler Thromb Vasc Biol. 2012 Nov;32(11):2541-6. doi: 10.1161/ATVBAHA.112.250571. Epub 2012 Aug 30. Arterioscler Thromb Vasc Biol. 2012. PMID: 22936343 Free PMC article. Review.
Cited by
-
Protective Effects of Rat Bone Marrow Mesenchymal Stem Cells-Derived Fusogenic Plasma Membrane Vesicles Containing VSVG Protein Mediated Mitochondrial Transfer on Myocardial Injury In Vitro.FASEB Bioadv. 2025 Apr 15;7(5):e70010. doi: 10.1096/fba.2024-00235. eCollection 2025 May. FASEB Bioadv. 2025. PMID: 40330432 Free PMC article.
References
-
- Moreira A.S., Cavaco D.G., Faria T.Q., Alves P.M., Carrondo M.J.T., Peixoto C. Advances in Lentivirus Purification. Biotechnol. J. 2021;16 - PubMed
-
- Palfi S., Gurruchaga J.M., Ralph G.S., Lepetit H., Lavisse S., Buttery P.C., Watts C., Miskin J., Kelleher M., Deeley S., et al. Long-term safety and tolerability of ProSavin, a lentiviral vector-based gene therapy for Parkinson's disease: a dose escalation, open-label, phase 1/2 trial. Lancet. 2014;383:1138–1146. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

