Evolutionary history influences the microbiomes of a female symbiotic reproductive organ in cephalopods
- PMID: 38315021
- PMCID: PMC10952459
- DOI: 10.1128/aem.00990-23
Evolutionary history influences the microbiomes of a female symbiotic reproductive organ in cephalopods
Abstract
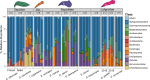
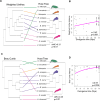
Many female squids and cuttlefishes have a symbiotic reproductive organ called the accessory nidamental gland (ANG) that hosts a bacterial consortium involved with egg defense against pathogens and fouling organisms. While the ANG is found in multiple cephalopod families, little is known about the global microbial diversity of these ANG bacterial symbionts. We used 16S rRNA gene community analysis to characterize the ANG microbiome from different cephalopod species and assess the relationship between host and symbiont phylogenies. The ANG microbiome of 11 species of cephalopods from four families (superorder: Decapodiformes) that span seven geographic locations was characterized. Bacteria of class Alphaproteobacteria, Gammaproteobacteria, and Flavobacteriia were found in all species, yet analysis of amplicon sequence variants by multiple distance metrics revealed a significant difference between ANG microbiomes of cephalopod families (weighted/unweighted UniFrac, Bray-Curtis, P = 0.001). Despite being collected from widely disparate geographic locations, members of the family Sepiolidae (bobtail squid) shared many bacterial taxa including (~50%) Opitutae (Verrucomicrobia) and Ruegeria (Alphaproteobacteria) species. Furthermore, we tested for phylosymbiosis and found a positive correlation between host phylogenetic distance and bacterial community dissimilarity (Mantel test r = 0.7). These data suggest that closely related sepiolids select for distinct symbionts from similar bacterial taxa. Overall, the ANGs of different cephalopod species harbor distinct microbiomes and thus offer a diverse symbiont community to explore antimicrobial activity and other functional roles in host fitness.IMPORTANCEMany aquatic organisms recruit microbial symbionts from the environment that provide a variety of functions, including defense from pathogens. Some female cephalopods (squids, bobtail squids, and cuttlefish) have a reproductive organ called the accessory nidamental gland (ANG) that contains a bacterial consortium that protects eggs from pathogens. Despite the wide distribution of these cephalopods, whether they share similar microbiomes is unknown. Here, we studied the microbial diversity of the ANG in 11 species of cephalopods distributed over a broad geographic range and representing 15-120 million years of host divergence. The ANG microbiomes shared some bacterial taxa, but each cephalopod species had unique symbiotic members. Additionally, analysis of host-symbiont phylogenies suggests that the evolutionary histories of the partners have been important in shaping the ANG microbiome. This study advances our knowledge of cephalopod-bacteria relationships and provides a foundation to explore defensive symbionts in other systems.
Keywords: Alphaproteobacteria; Verrucomicrobia; accessory nidamental gland; cephalopod; microbial communities; microbiomes; phylosymbiosis; symbiosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Lum‐Kong A, Hastings TS. 1992. The accessory nidamental glands of Loligo forbesi (cephalopoda: loliginidae): characterization of symbiotic bacteria and preliminary experiments to investigate factors controlling sexual maturation. J Zoology 228:395–403. doi:10.1111/j.1469-7998.1992.tb04443.x - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

