Genetic diversity and recombination of bovine enterovirus strains in China
- PMID: 38315051
- PMCID: PMC10913430
- DOI: 10.1128/spectrum.02800-23
Genetic diversity and recombination of bovine enterovirus strains in China
Abstract
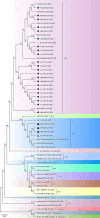
Bovine enterovirus (BEV) consisting of enterovirus species E (EV-E) and F (EV-F) is the causative agent associated with respiratory and gastrointestinal diseases in cattle. Here, we reported the characterization, genetic diversity, and recombination of novel BEV strains isolated from the major cattle-raising regions in China during 2012-2018. Twenty-seven BEV strains were successfully isolated and characterized. Molecular characterization demonstrated that the majority of these novel BEV strains (24/27) were EV-E, while only few strains (3/27) were EV-F. Sequence analysis revealed the diversity of the circulating BEV strains such as species and subtypes where different species or subtype coinfections were detected in the same regions and even in the same cattle herds. For the EV-E, two novel subtypes, designated as EV-E6 and EV-E7, were revealed in addition to the currently reported EV-E1-EV-E5. Comparative genomic analysis revealed the intraspecies and interspecies genetic exchanges among BEV isolates. The representative strain HeN-B62 was probably from AN12 (EV-F7) and PS-87-Belfast (EV-F3) strains. The interspecies recombination between EV-E and EV-F was also discovered, where the EV-F7-AN12 might be from EV-E5 and EV-F1, and EV-E5-MexKSU/5 may be recombined from EV-F7 and EV-E1. The aforementioned results revealed the genetic diversity and recombination of novel BEV strains and unveiled the different BEV species or subtype infections in the same cattle herd, which will broaden the understanding of enterovirus genetic diversity, recombination, pathogenesis, and prevention of disease outbreaks.
Importance: Bovine enterovirus (BEV) infection is an emerging disease in China that is characterized by digestive, respiratory, and reproductive disorders. In this study, we first reported two novel EV-E subtypes detected in cattle herds in China, unveiled the coinfection of two enterovirus species (EV-E/EV-F) and different subtypes (EV-E2/EV-E7, EV-E1/EV-E7, and EV-E3/EV-E6) in the same cattle herds, and revealed the enterovirus genetic exchange in intraspecies and interspecies recombination. These results provide an important update of enterovirus prevalence and epidemiological aspects and contribute to a better understanding of enterovirus genetic diversity, evolution, and pathogenesis.
Keywords: bovine enterovirus; cattle; isolation; molecular analysis; recombination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Isolation and Identification of Two Clinical Strains of the Novel Genotype Enterovirus E5 in China.Microbiol Spectr. 2022 Jun 29;10(3):e0266221. doi: 10.1128/spectrum.02662-21. Epub 2022 Jun 2. Microbiol Spectr. 2022. PMID: 35652637 Free PMC article.
-
Molecular Epidemiology of Bovine Enteroviruses and Genome Characterization of Two Novel Bovine Enterovirus Strains in Guangxi, China.Microbiol Spectr. 2023 Mar 6;11(2):e0378522. doi: 10.1128/spectrum.03785-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36877012 Free PMC article.
-
Identification of a novel bovine enterovirus possessing highly divergent amino acid sequences in capsid protein.BMC Microbiol. 2017 Jan 17;17(1):18. doi: 10.1186/s12866-016-0923-0. BMC Microbiol. 2017. PMID: 28095784 Free PMC article.
-
Bovine enterovirus 2: complete genomic sequence and molecular modelling of a reference strain and a wild-type isolate from endemically infected US cattle.J Gen Virol. 2004 Nov;85(Pt 11):3195-3203. doi: 10.1099/vir.0.80159-0. J Gen Virol. 2004. PMID: 15483232
-
Circulation of two Enterovirus C105 (EV-C105) lineages in Europe and Africa.J Gen Virol. 2015 Jun;96(Pt 6):1374-1379. doi: 10.1099/vir.0.000088. Epub 2015 Feb 9. J Gen Virol. 2015. PMID: 25667329 Review.
Cited by
-
Genomic Characterization of Two Bovine Enterovirus Strains Isolated from Newly Transported Cattle.Vet Sci. 2025 Jul 11;12(7):660. doi: 10.3390/vetsci12070660. Vet Sci. 2025. PMID: 40711320 Free PMC article.
-
Enterovirus E infections in goats with respiratory disease.BMC Vet Res. 2025 Feb 17;21(1):71. doi: 10.1186/s12917-025-04537-x. BMC Vet Res. 2025. PMID: 39962514 Free PMC article.
References
-
- Zerbini FM, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, Dempsey DM, Dutilh BE, García ML, Hendrickson RC, Junglen S, Krupovic M, Kuhn JH, Lambert AJ, Łobocka M, Oksanen HM, Robertson DL, Rubino L, Sabanadzovic S, Simmonds P, Smith DB, Suzuki N, Van Doorslaer K, Vandamme A-M, Varsani A. 2023. Changes to virus taxonomy and the ICTV statutes ratified by the International Committee on Taxonomy of viruses. Arch Virol 168:175. doi:10.1007/s00705-023-05797-4 - DOI - PMC - PubMed
-
- Zell R, Delwart E, Gorbalenya AE, Hovi T, King AMQ, Knowles NJ, Lindberg AM, Pallansch MA, Palmenberg AC, Reuter G, Simmonds P, Skern T, Stanway G, Yamashita TIctv Report Consortium . 2017. ICTV virus taxonomy profile: picornaviridae. J Gen Virol 98:2421–2422. doi:10.1099/jgv.0.000911 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

