Compensatory mutations are associated with increased in vitro growth in resistant clinical samples of Mycobacterium tuberculosis
- PMID: 38315172
- PMCID: PMC10926696
- DOI: 10.1099/mgen.0.001187
Compensatory mutations are associated with increased in vitro growth in resistant clinical samples of Mycobacterium tuberculosis
Abstract
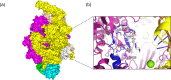
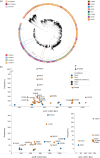
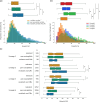
Mutations in Mycobacterium tuberculosis associated with resistance to antibiotics often come with a fitness cost for the bacteria. Resistance to the first-line drug rifampicin leads to lower competitive fitness of M. tuberculosis populations when compared to susceptible populations. This fitness cost, introduced by resistance mutations in the RNA polymerase, can be alleviated by compensatory mutations (CMs) in other regions of the affected protein. CMs are of particular interest clinically since they could lock in resistance mutations, encouraging the spread of resistant strains worldwide. Here, we report the statistical inference of a comprehensive set of CMs in the RNA polymerase of M. tuberculosis, using over 70 000 M. tuberculosis genomes that were collated as part of the CRyPTIC project. The unprecedented size of this data set gave the statistical tests more power to investigate the association of putative CMs with resistance-conferring mutations. Overall, we propose 51 high-confidence CMs by means of statistical association testing and suggest hypotheses for how they exert their compensatory mechanism by mapping them onto the protein structure. In addition, we were able to show an association of CMs with higher in vitro growth densities, and hence presumably with higher fitness, in resistant samples in the more virulent M. tuberculosis lineage 2. Our results suggest the association of CM presence with significantly higher in vitro growth than for wild-type samples, although this association is confounded with lineage and sub-lineage affiliation. Our findings emphasize the integral role of CMs and lineage affiliation in resistance spread and increases the urgency of antibiotic stewardship, which implies accurate, cheap and widely accessible diagnostics for M. tuberculosis infections to not only improve patient outcomes but also prevent the spread of resistant strains.
Keywords: antimicrobial resistance; compensatory mutations; fitness cost; tuberculosis.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



Similar articles
-
Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes.Nat Genet. 2011 Dec 18;44(1):106-10. doi: 10.1038/ng.1038. Nat Genet. 2011. PMID: 22179134 Free PMC article.
-
Putative compensatory mutations in the rpoC gene of rifampin-resistant Mycobacterium tuberculosis are associated with ongoing transmission.Antimicrob Agents Chemother. 2013 Feb;57(2):827-32. doi: 10.1128/AAC.01541-12. Epub 2012 Dec 3. Antimicrob Agents Chemother. 2013. PMID: 23208709 Free PMC article.
-
Compensatory effects of M. tuberculosis rpoB mutations outside the rifampicin resistance-determining region.Emerg Microbes Infect. 2021 Dec;10(1):743-752. doi: 10.1080/22221751.2021.1908096. Emerg Microbes Infect. 2021. PMID: 33775224 Free PMC article.
-
Evolution of Mycobacterium tuberculosis drug resistance in the genomic era.Front Cell Infect Microbiol. 2022 Oct 7;12:954074. doi: 10.3389/fcimb.2022.954074. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36275027 Free PMC article. Review.
-
Drug resistance, fitness and compensatory mutations in Mycobacterium tuberculosis.Tuberculosis (Edinb). 2021 Jul;129:102091. doi: 10.1016/j.tube.2021.102091. Epub 2021 May 21. Tuberculosis (Edinb). 2021. PMID: 34090078 Review.
Cited by
-
Rifampicin and isoniazid resistance not promote fluoroquinolone resistance in Mycobacterium smegmatis.PLoS One. 2025 Jan 2;20(1):e0315512. doi: 10.1371/journal.pone.0315512. eCollection 2025. PLoS One. 2025. PMID: 39746086 Free PMC article.
-
Multidrug-resistant tuberculosis clusters and transmission in Taiwan: a population-based cohort study.Front Microbiol. 2024 Sep 18;15:1439532. doi: 10.3389/fmicb.2024.1439532. eCollection 2024. Front Microbiol. 2024. PMID: 39360329 Free PMC article.
-
Ecology, global diversity and evolutionary mechanisms in the Mycobacterium tuberculosis complex.Nat Rev Microbiol. 2025 Sep;23(9):602-614. doi: 10.1038/s41579-025-01159-w. Epub 2025 Mar 25. Nat Rev Microbiol. 2025. PMID: 40133503 Review.
-
Multi-drug resistance and compensatory mutations in Mycobacterium tuberculosis in Vietnam.Trop Med Int Health. 2025 May;30(5):426-436. doi: 10.1111/tmi.14104. Epub 2025 Mar 13. Trop Med Int Health. 2025. PMID: 40078052 Free PMC article.
-
Predicting rifampicin resistance in Mycobacterium tuberculosis using machine learning informed by protein structural and chemical features.ERJ Open Res. 2025 Jun 30;11(3):00952-2024. doi: 10.1183/23120541.00952-2024. eCollection 2025 May. ERJ Open Res. 2025. PMID: 40589903 Free PMC article.
References
-
- Brunner V, Fowler PW. Accompanying code to reproduce analysis and figures. https://github.com/fowler-lab/tb-rnap-compensation n.d.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

