CRISPR screen for protein inclusion formation uncovers a role for SRRD in the regulation of intermediate filament dynamics and aggresome assembly
- PMID: 38315730
- PMCID: PMC10868785
- DOI: 10.1371/journal.pgen.1011138
CRISPR screen for protein inclusion formation uncovers a role for SRRD in the regulation of intermediate filament dynamics and aggresome assembly
Abstract
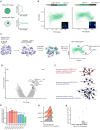
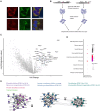
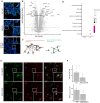
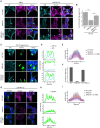
The presence of large protein inclusions is a hallmark of neurodegeneration, and yet the precise molecular factors that contribute to their formation remain poorly understood. Screens using aggregation-prone proteins have commonly relied on downstream toxicity as a readout rather than the direct formation of aggregates. Here, we combined a genome-wide CRISPR knockout screen with Pulse Shape Analysis, a FACS-based method for inclusion detection, to identify direct modifiers of TDP-43 aggregation in human cells. Our screen revealed both canonical and novel proteostasis genes, and unearthed SRRD, a poorly characterized protein, as a top regulator of protein inclusion formation. APEX biotin labeling reveals that SRRD resides in proximity to proteins that are involved in the formation and breakage of disulfide bonds and to intermediate filaments, suggesting a role in regulation of the spatial dynamics of the intermediate filament network. Indeed, loss of SRRD results in aberrant intermediate filament fibrils and the impaired formation of aggresomes, including blunted vimentin cage structure, during proteotoxic stress. Interestingly, SRRD also localizes to aggresomes and unfolded proteins, and rescues proteotoxicity in yeast whereby its N-terminal low complexity domain is sufficient to induce this affect. Altogether this suggests an unanticipated and broad role for SRRD in cytoskeletal organization and cellular proteostasis.
Copyright: © 2024 Sweeney et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: OS and KMS have filed a patent application for the use of SRRD fragments through the Children’s Hospital of Philadelphia. J.S. is a consultant for Dewpoint Therapeutics, ADRx, and Neumora. J.S. is a shareholder and advisor for Confluence Therapeutics. The authors declare no other conflicts of interest relevant to this publication.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

