Integrating AI-Powered Digital Pathology and Imaging Mass Cytometry Identifies Key Classifiers of Tumor Cells, Stroma, and Immune Cells in Non-Small Cell Lung Cancer
- PMID: 38315789
- PMCID: PMC10982643
- DOI: 10.1158/0008-5472.CAN-23-1698
Integrating AI-Powered Digital Pathology and Imaging Mass Cytometry Identifies Key Classifiers of Tumor Cells, Stroma, and Immune Cells in Non-Small Cell Lung Cancer
Abstract
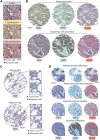
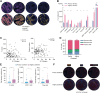
Artificial intelligence (AI)-powered approaches are becoming increasingly used as histopathologic tools to extract subvisual features and improve diagnostic workflows. On the other hand, hi-plex approaches are widely adopted to analyze the immune ecosystem in tumor specimens. Here, we aimed at combining AI-aided histopathology and imaging mass cytometry (IMC) to analyze the ecosystem of non-small cell lung cancer (NSCLC). An AI-based approach was used on hematoxylin and eosin (H&E) sections from 158 NSCLC specimens to accurately identify tumor cells, both adenocarcinoma and squamous carcinoma cells, and to generate a classifier of tumor cell spatial clustering. Consecutive tissue sections were stained with metal-labeled antibodies and processed through the IMC workflow, allowing quantitative detection of 24 markers related to tumor cells, tissue architecture, CD45+ myeloid and lymphoid cells, and immune activation. IMC identified 11 macrophage clusters that mainly localized in the stroma, except for S100A8+ cells, which infiltrated tumor nests. T cells were preferentially localized in peritumor areas or in tumor nests, the latter being associated with better prognosis, and they were more abundant in highly clustered tumors. Integrated tumor and immune classifiers were validated as prognostic on whole slides. In conclusion, integration of AI-powered H&E and multiparametric IMC allows investigation of spatial patterns and reveals tissue relevant features with clinical relevance.
Significance: Leveraging artificial intelligence-powered H&E analysis integrated with hi-plex imaging mass cytometry provides insights into the tumor ecosystem and can translate tumor features into classifiers to predict prognosis, genotype, and therapy response.
©2024 The Authors; Published by the American Association for Cancer Research.
Figures



References
-
- Kleppe A, Skrede OJ, De Raedt S, Liestøl K, Kerr DJ, Danielsen HE. Designing deep learning studies in cancer diagnostics. Nat Rev Cancer 2021;21:199–211. - PubMed
-
- Shmatko A, Ghaffari Laleh N, Gerstung M, Kather JN. Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nat Cancer 2022;3:1026–38. - PubMed
-
- Guramare M, Ashar Javed S, Agrawal N, Abel J, Montalto M, Beck A, et al. . Digital pathology uncovers multi-omic hallmarks of lung cancer in histopathology images. J Thorac Oncol 2023;17:15–15.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

