G-quadruplex DNA structure is a positive regulator of MYC transcription
- PMID: 38315865
- PMCID: PMC10873556
- DOI: 10.1073/pnas.2320240121
G-quadruplex DNA structure is a positive regulator of MYC transcription
Abstract
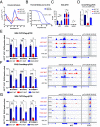
DNA structure can regulate genome function. Four-stranded DNA G-quadruplex (G4) structures have been implicated in transcriptional regulation; however, previous studies have not directly addressed the role of an individual G4 within its endogenous cellular context. Using CRISPR to genetically abrogate endogenous G4 structure folding, we directly interrogate the G4 found within the upstream regulatory region of the critical human MYC oncogene. G4 loss leads to suppression of MYC transcription from the P1 promoter that is mediated by the deposition of a de novo nucleosome alongside alterations in RNA polymerase recruitment. We also show that replacement of the endogenous MYC G4 with a different G4 structure from the KRAS oncogene restores G4 folding and MYC transcription. Moreover, we demonstrate that the MYC G4 structure itself, rather than its sequence, recruits transcription factors and histone modifiers. Overall, our work establishes that G4 structures are important features of transcriptional regulation that coordinate recruitment of key chromatin proteins and the transcriptional machinery through interactions with DNA secondary structure, rather than primary sequence.
Keywords: DNA; G-quadruplex; MYC; epigenetics; transcription.
Conflict of interest statement
Competing interests statement:S.B. is a founder, shareholder, and paid advisor and Director of Biomodal Ltd. and Inflex Ltd.; a founder, shareholder, and director of Elyx Ltd.; a founder and shareholder of RNAvate Ltd. L.M. is a paid consultant for Inflex Ltd. S.B. is a founder and shareholder (>5%) of Biomodal Ltd., Inflex Ltd., Elyx Ltd., and RNAvate Ltd.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

