Accelerated Lung Function Decline and Mucus-Microbe Evolution in Chronic Obstructive Pulmonary Disease
- PMID: 38315959
- PMCID: PMC11348959
- DOI: 10.1164/rccm.202306-1060OC
Accelerated Lung Function Decline and Mucus-Microbe Evolution in Chronic Obstructive Pulmonary Disease
Abstract
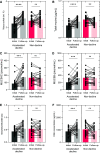
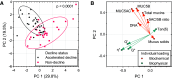
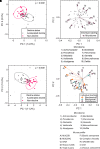
Rationale: Progressive lung function loss is recognized in chronic obstructive pulmonary disease (COPD); however, no study concurrently evaluates how accelerated lung function decline relates to mucus properties and the microbiome in COPD. Objectives: Longitudinal assessment of mucus and microbiome changes accompanying accelerated lung function decline in patients COPD. Methods: This was a prospective, longitudinal assessment of the London COPD cohort exhibiting the greatest FEV1 decline (n = 30; accelerated decline; 156 ml/yr FEV1 loss) and with no FEV1 decline (n = 28; nondecline; 49 ml/yr FEV1 gain) over time. Lung microbiomes from paired sputum (total 116 specimens) were assessed by shotgun metagenomics and corresponding mucus profiles evaluated for biochemical and biophysical properties. Measurements and Main Results: Biochemical and biophysical mucus properties are significantly altered in the accelerated decline group. Unsupervised principal component analysis showed clear separation, with mucus biochemistry associated with accelerated decline, whereas biophysical mucus characteristics contributed to interindividual variability. When mucus and microbes are considered together, an accelerated decline mucus-microbiome association emerges, characterized by increased mucin (MUC5AC [mucin 5AC] and MUC5B [mucin 5B]) concentration and the presence of Achromobacter and Klebsiella. As COPD progresses, mucus-microbiome shifts occur, initially characterized by low mucin concentration and transition from viscous to elastic dominance accompanied by the commensals Veillonella, Gemella, Rothia, and Prevotella (Global Initiative for Chronic Obstructive Lung Disease [GOLD] A and B) before transition to increased mucus viscosity, mucins, and DNA concentration together with the emergence of pathogenic microorganisms including Haemophilus, Moraxella, and Pseudomonas (GOLD E). Conclusions: Mucus-microbiome associations evolve over time with accelerated lung function decline, symptom progression, and exacerbations affording fresh therapeutic opportunities for early intervention.
Keywords: COPD; lung function decline; metagenomics; mucus; rheology.
Figures


Comment in
-
Of Mucus and Microbes: The Sticky Issue of Mucin-Microbiome Interactions in Chronic Obstructive Pulmonary Disease.Am J Respir Crit Care Med. 2024 Aug 1;210(3):252-253. doi: 10.1164/rccm.202403-0506ED. Am J Respir Crit Care Med. 2024. PMID: 38530107 Free PMC article. No abstract available.
References
-
- Yang IA, Jenkins CR, Salvi SS. Chronic obstructive pulmonary disease in never-smokers: risk factors, pathogenesis, and implications for prevention and treatment. Lancet Respir Med . 2022;10:497–511. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 020458-00001/LKCMedicine-ICL Fellowship
- LEARN008/LKCMedicine LEARN Grant
- SGL026/1079/National Heart and Lung Institute (NHLI) Lectureship
- MOH-000141/National Medical Research Council
- MOH-001356/National Medical Research Council
- MOH-000710/National Medical Research Council
- RT1/22/Singapore Ministry of Education under its AcRF Tier 1 Grant
- National Institute of Health Research (NIHR) Biomedical Research Centre to Imperial College London
- National Institute of Health Research (NIHR) Biomedical Research Centre to Royal Brompton Hospital
LinkOut - more resources
Full Text Sources
Medical
Research Materials

