Stability of gut microbiome after COVID-19 vaccination in healthy and immuno-compromised individuals
- PMID: 38316462
- PMCID: PMC10844540
- DOI: 10.26508/lsa.202302529
Stability of gut microbiome after COVID-19 vaccination in healthy and immuno-compromised individuals
Abstract
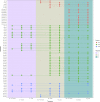
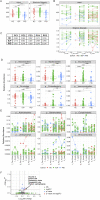
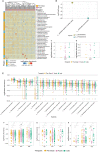
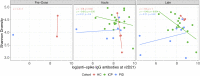
Bidirectional interactions between the immune system and the gut microbiota are key contributors to various physiological functions. Immune-associated diseases such as cancer and autoimmunity, and efficacy of immunomodulatory therapies, have been linked to microbiome variation. Although COVID-19 infection has been shown to cause microbial dysbiosis, it remains understudied whether the inflammatory response associated with vaccination also impacts the microbiota. Here, we investigate the temporal impact of COVID-19 vaccination on the gut microbiome in healthy and immuno-compromised individuals; the latter included patients with primary immunodeficiency and cancer patients on immunomodulating therapies. We find that the gut microbiome remained remarkably stable post-vaccination irrespective of diverse immune status, vaccine response, and microbial composition spanned by the cohort. The stability is evident at all evaluated levels including diversity, phylum, species, and functional capacity. Our results indicate the resilience of the gut microbiome to host immune changes triggered by COVID-19 vaccination and suggest minimal, if any, impact on microbiome-mediated processes. These findings encourage vaccine acceptance, particularly when contrasted with the significant microbiome shifts observed during COVID-19 infection.
© 2024 Boston et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- WHO (2023) WHO Coronavirus (COVID-19) Dashboard. Available at: https://covid19.who.int
-
- Lind ML, Dorion M, Houde AJ, Lansing M, Lapidus S, Thomas R, Yildirim I, Omer SB, Schulz WL, Andrews JR, et al. (2023) Evidence of leaky protection following COVID-19 vaccination and SARS-CoV-2 infection in an incarcerated population. Nat Commun 14: 5055. 10.1038/s41467-023-40750-8 - DOI - PMC - PubMed
-
- Bergamaschi C, Terpos E, Rosati M, Angel M, Bear J, Stellas D, Karaliota S, Apostolakou F, Bagratuni T, Patseas D, et al. (2021) Systemic IL-15, IFN-gamma, and IP-10/CXCL10 signature associated with effective immune response to SARS-CoV-2 in BNT162b2 mRNA vaccine recipients. Cell Rep 36: 109504. 10.1016/j.celrep.2021.109504 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
