Single-cell insights into immune dysregulation in rheumatoid arthritis flare versus drug-free remission
- PMID: 38316770
- PMCID: PMC10844292
- DOI: 10.1038/s41467-024-45213-2
Single-cell insights into immune dysregulation in rheumatoid arthritis flare versus drug-free remission
Abstract
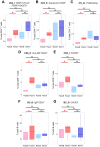
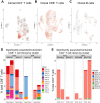
Immune-mediated inflammatory diseases (IMIDs) are typically characterised by relapsing and remitting flares of inflammation. However, the unpredictability of disease flares impedes their study. Addressing this critical knowledge gap, we use the experimental medicine approach of immunomodulatory drug withdrawal in rheumatoid arthritis (RA) remission to synchronise flare processes allowing detailed characterisation. Exploratory mass cytometry analyses reveal three circulating cellular subsets heralding the onset of arthritis flare - CD45RO+PD1hi CD4+ and CD8+ T cells, and CD27+CD86+CD21- B cells - further characterised by single-cell sequencing. Distinct lymphocyte subsets including cytotoxic and exhausted CD4+ memory T cells, memory CD8+CXCR5+ T cells, and IGHA1+ plasma cells are primed for activation in flare patients. Regulatory memory CD4+ T cells (Treg cells) increase at flare onset, but with dysfunctional regulatory marker expression compared to drug-free remission. Significant clonal expansion is observed in T cells, but not B cells, after drug cessation; this is widespread throughout memory CD8+ T cell subsets but limited to the granzyme-expressing cytotoxic subset within CD4+ memory T cells. Based on our observations, we suggest a model of immune dysregulation for understanding RA flare, with potential for further translational research towards novel avenues for its treatment and prevention.
© 2024. The Author(s).
Conflict of interest statement
K.F.B., A.G.P. and J.D.I. are named as inventors on a patent application by Newcastle University (“Prediction of Drug-Free Remission in Rheumatoid Arthritis”; International Patent Application Number PCT/GB2019/050902). D.Mc.D., G.H., R.H., J.C., D.S., A.R.S., H.E.M., L.Mac.D., A.F. and A.E.A. have no competing interests to declare.
Figures




References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

