Comparative analyses of Netherton syndrome patients and Spink5 conditional knock-out mice uncover disease-relevant pathways
- PMID: 38316920
- PMCID: PMC10844249
- DOI: 10.1038/s42003-024-05780-y
Comparative analyses of Netherton syndrome patients and Spink5 conditional knock-out mice uncover disease-relevant pathways
Abstract
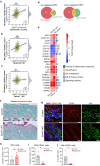
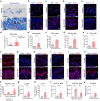
Netherton syndrome (NS) is a rare skin disease caused by loss-of-function mutations in the serine peptidase inhibitor Kazal type 5 (SPINK5) gene. Disease severity and the lack of efficacious treatments call for a better understanding of NS mechanisms. Here we describe a novel and viable, Spink5 conditional knock-out (cKO) mouse model, allowing to study NS progression. By combining transcriptomics and proteomics, we determine a disease molecular profile common to mouse models and NS patients. Spink5 cKO mice and NS patients share skin barrier and inflammation signatures defined by up-regulation and increased activity of proteases, IL-17, IL-36, and IL-20 family cytokine signaling. Systemic inflammation in Spink5 cKO mice correlates with disease severity and is associated with thymic atrophy and enlargement of lymph nodes and spleen. This systemic inflammation phenotype is marked by neutrophils and IL-17/IL-22 signaling, does not involve primary T cell immunodeficiency and is independent of bacterial infection. By comparing skin transcriptomes and proteomes, we uncover several putative substrates of tissue kallikrein-related proteases (KLKs), demonstrating that KLKs can proteolytically regulate IL-36 pro-inflammatory cytokines. Our study thus provides a conserved molecular framework for NS and reveals a KLK/IL-36 signaling axis, adding new insights into the disease mechanisms and therapeutic targets.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interest(s): E.P., C.B. and A.H. are inventors of a patent. The rest of the authors declare no competing interests.
Figures






References
-
- Petrova E, Hovnanian A. Advances in understanding of Netherton syndrome and therapeutic implications. Expert Opin. Orphan Drugs. 2020;8:455–487. doi: 10.1080/21678707.2020.1857724. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

