Sterile inflammation via TRPM8 RNA-dependent TLR3-NF-kB/IRF3 activation promotes antitumor immunity in prostate cancer
- PMID: 38316991
- PMCID: PMC10907604
- DOI: 10.1038/s44318-024-00040-5
Sterile inflammation via TRPM8 RNA-dependent TLR3-NF-kB/IRF3 activation promotes antitumor immunity in prostate cancer
Abstract
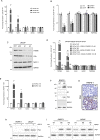
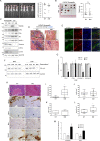
Inflammation is a common condition of prostate tissue, whose impact on carcinogenesis is highly debated. Microbial colonization is a well-documented cause of a small percentage of prostatitis cases, but it remains unclear what underlies the majority of sterile inflammation reported. Here, androgen- independent fluctuations of PSA expression in prostate cells have lead us to identify a prominent function of the Transient Receptor Potential Cation Channel Subfamily M Member 8 (TRPM8) gene in sterile inflammation. Prostate cells secret TRPM8 RNA into extracellular vesicles (EVs), which primes TLR3/NF-kB-mediated inflammatory signaling after EV endocytosis by epithelial cancer cells. Furthermore, prostate cancer xenografts expressing a translation-defective form of TRPM8 RNA contain less collagen type I in the extracellular matrix, significantly more infiltrating NK cells, and larger necrotic areas as compared to control xenografts. These findings imply sustained, androgen-independent expression of TRPM8 constitutes as a promoter of anticancer innate immunity, which may constitute a clinically relevant condition affecting prostate cancer prognosis.
Keywords: Immunity; Inflammation; PSA; Prostate; TRP.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Alaimo A, Lorenzoni M, Ambrosino P, Bertossi A, Bisio A, Macchia A, Zoni E, Genovesi S, Cambuli F, Foletto V, et al. Calcium cytotoxicity sensitizes prostate cancer cells to standard-of-care treatments for locally advanced tumors. Cell Death Dis. 2020;11:1039. doi: 10.1038/s41419-020-03256-5. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

