Historic methicillin-resistant Staphylococcus aureus: expanding current knowledge using molecular epidemiological characterization of a Swiss legacy collection
- PMID: 38317199
- PMCID: PMC10840241
- DOI: 10.1186/s13073-024-01292-w
Historic methicillin-resistant Staphylococcus aureus: expanding current knowledge using molecular epidemiological characterization of a Swiss legacy collection
Abstract
Background: Few methicillin-resistant Staphylococcus aureus (MRSA) from the early years of its global emergence have been sequenced. Knowledge about evolutionary factors promoting the success of specific MRSA multi-locus sequence types (MLSTs) remains scarce. We aimed to characterize a legacy MRSA collection isolated from 1965 to 1987 and compare it against publicly available international and local genomes.
Methods: We accessed 451 historic (1965-1987) MRSA isolates stored in the Culture Collection of Switzerland, mostly collected from the Zurich region. We determined phenotypic antimicrobial resistance (AMR) and performed whole genome sequencing (WGS) using Illumina short-read sequencing on all isolates and long-read sequencing on a selection with Oxford Nanopore Technology. For context, we included 103 publicly available international assemblies from 1960 to 1992 and sequenced 1207 modern Swiss MRSA isolates from 2007 to 2022. We analyzed the core genome (cg)MLST and predicted SCCmec cassette types, AMR, and virulence genes.
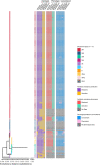
Results: Among the 451 historic Swiss MRSA isolates, we found 17 sequence types (STs) of which 11 have been previously described. Two STs were novel combinations of known loci and six isolates carried previously unsubmitted MLST alleles, representing five new STs (ST7843, ST7844, ST7837, ST7839, and ST7842). Most isolates (83% 376/451) represented ST247-MRSA-I isolated in the 1960s, followed by ST7844 (6% 25/451), a novel single locus variant (SLV) of ST239. Analysis by cgMLST indicated that isolates belonging to ST7844-MRSA-III cluster within the diversity of ST239-MRSA-III. Early MRSA were predominantly from clonal complex (CC)8. From 1980 to the end of the twentieth century, we observed that CC22 and CC5 as well as CC8 were present, both locally and internationally.
Conclusions: The combined analysis of 1761 historic and contemporary MRSA isolates across more than 50 years uncovered novel STs and allowed us a glimpse into the lineage flux between Swiss-German and international MRSA across time.
Keywords: Antimicrobial resistance; Historic; MLST; MRSA; Phylogeny; Switzerland; WGS.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
[Infectivity-resistotype-genotype clustering of methicillin-resistant Staphylococcus aureus strains in the Central Blacksea Region of Turkey].Mikrobiyol Bul. 2014 Jan;48(1):14-27. Mikrobiyol Bul. 2014. PMID: 24506712 Turkish.
-
Molecular epidemiology of Staphylococcus aureus bacteremia in a single large Minnesota medical center in 2015 as assessed using MLST, core genome MLST and spa typing.PLoS One. 2017 Jun 2;12(6):e0179003. doi: 10.1371/journal.pone.0179003. eCollection 2017. PLoS One. 2017. PMID: 28575112 Free PMC article.
-
Drivers of methicillin-resistant Staphylococcus aureus (MRSA) lineage replacement in China.Genome Med. 2021 Oct 28;13(1):171. doi: 10.1186/s13073-021-00992-x. Genome Med. 2021. PMID: 34711267 Free PMC article.
-
Emergence of CC8/ST239- SCCmec III/t421 tigecycline resistant and CC/ST22-SCCmec IV/t790 vancomycin resistant Staphylococcus aureus strains isolated from wound: A two-year multi-center study in Tehran, Iran.Acta Microbiol Immunol Hung. 2021 Nov 22;68(4):227-234. doi: 10.1556/030.2021.01534. Print 2021 Dec 2. Acta Microbiol Immunol Hung. 2021. PMID: 34806999
-
Molecular epidemiology and antibiotic resistance of methicillin-resistant Staphylococcus aureus circulating in the Russian Federation.Infect Genet Evol. 2017 Sep;53:189-194. doi: 10.1016/j.meegid.2017.06.006. Epub 2017 Jun 7. Infect Genet Evol. 2017. PMID: 28600216
Cited by
-
Genomics for antimicrobial resistance-progress and future directions.Antimicrob Agents Chemother. 2025 May 7;69(5):e0108224. doi: 10.1128/aac.01082-24. Epub 2025 Apr 14. Antimicrob Agents Chemother. 2025. PMID: 40227048 Free PMC article. Review.
References
-
- Rammelkamp CH, Maxon T. Resistance of staphylococcus aureus to the action of penicillin. Proc Soc Exp Biol Med. 1942;51(3):386–9. doi: 10.3181/00379727-51-13986. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

