Exploring the DNA2-PNA heterotriplex formation in targeting the Bcl-2 gene promoter: A structural insight by physico-chemical and microsecond-scale MD investigation
- PMID: 38317891
- PMCID: PMC10839560
- DOI: 10.1016/j.heliyon.2024.e24599
Exploring the DNA2-PNA heterotriplex formation in targeting the Bcl-2 gene promoter: A structural insight by physico-chemical and microsecond-scale MD investigation
Abstract
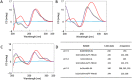
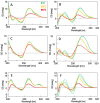
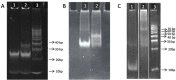
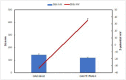
Peptide Nucleic Acids (PNAs) represent a promising tool for gene modulation in anticancer treatment. The uncharged peptidyl backbone and the resistance to chemical and enzymatic degradation make PNAs highly advantageous to form stable hybrid complexes with complementary DNA and RNA strands, providing higher stability than the corresponding natural analogues. Our and other groups' research has successfully shown that tailored PNA sequences can effectively downregulate the expression of human oncogenes using antigene, antisense, or anti-miRNA approaches. Specifically, we identified a seven bases-long PNA sequence, complementary to the longer loop of the main G-quadruplex structure formed by the bcl2midG4 promoter sequence, capable of downregulating the expression of the antiapoptotic Bcl-2 protein and enhancing the anticancer activity of an oncolytic adenovirus. Here, we extended the length of the PNA probe with the aim of including the double-stranded Bcl-2 promoter among the targets of the PNA probe. Our investigation primarily focused on the structural aspects of the resulting DNA2-PNA heterotriplex that were determined by employing conventional and accelerated microsecond-scale molecular dynamics simulations and chemical-physical analysis. Additionally, we conducted preliminary biological experiments using cytotoxicity assays on human A549 and MDA-MB-436 adenocarcinoma cell lines, employing the oncolytic adenovirus delivery strategy.
Keywords: Ad5Delta24 (OAd); Bcl-2 gene modulation; Drug delivery; PNA anti-Bcl-2; PNA gene expression modifiers; PNA-adenoviral vector.
© 2024 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Amato J., Pagano B., Borbone N., Oliviero G., Gabelica V., De Pauw E., D'Errico S., Piccialli V., Varra M., Giancola C., Piccialli G., Mayol L. Targeting G-quadruplex structure in the human c-kit promoter with short PNA sequences. Bioconjugate Chem. 2011;22:654–663. doi: 10.1021/bc100444v. - DOI - PubMed
-
- McNeer N.A., Schleifman E.B., Cuthbert A., Brehm M., Jackson A., Cheng C., Anandalingam K., Kumar P., Shultz L.D., Greiner D.L., Mark Saltzman W., Glazer P.M. Systemic delivery of triplex-forming PNA and donor DNA by nanoparticles mediates site-specific genome editing of human hematopoietic cells in vivo. Gene Ther. 2013;20:658–669. doi: 10.1038/gt.2012.82. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

