Compartmentalization and synergy of osteoblasts drive bone formation in the regenerating fin
- PMID: 38318374
- PMCID: PMC10838958
- DOI: 10.1016/j.isci.2024.108841
Compartmentalization and synergy of osteoblasts drive bone formation in the regenerating fin
Abstract
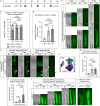
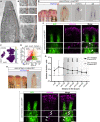
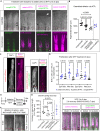
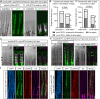
Zebrafish regenerate their fins which involves a component of cell plasticity. It is currently unclear how regenerate cells divide labor to allow for appropriate growth and patterning. Here, we studied lineage relationships of fluorescence-activated cell sorting-enriched epidermal, bone-forming (osteoblast), and (non-osteoblast) blastemal fin regenerate cells by single-cell RNA sequencing, lineage tracing, targeted osteoblast ablation, and electron microscopy. Most osteoblasts in the outgrowing regenerate derive from osterix+ osteoblasts, while mmp9+ cells reside at segment joints. Distal blastema cells contribute to distal osteoblast progenitors, suggesting compartmentalization of the regenerating appendage. Ablation of osterix+ osteoblasts impairs segment joint and bone matrix formation and decreases regenerate length which is partially compensated for by distal regenerate cells. Our study characterizes expression patterns and lineage relationships of rare fin regenerate cell populations, indicates inherent detection and compensation of impaired regeneration, suggests variable dependence on growth factor signaling, and demonstrates zonation of the elongating fin regenerate.
Keywords: Animal physiology; Biological sciences; Developmental biology; Natural sciences; Physiology.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Dietrich K., Fiedler I., Kurzyukova A., López-Delgado A.C., McGowan L., Geurtzen K., Hammond C.L., Busse B., Knopf F. Skeletal biology and disease modeling in zebrafish. J. Bone Miner. Res. 2021;36:436–458. - PubMed
-
- Sehring I.M., Weidinger G. Recent advancements in understanding fin regeneration in zebrafish. WIREs Dev. Biol. 2020;9:e367. - PubMed
-
- Nechiporuk A., Keating M.T. A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development. 2002;129:2607–2617. - PubMed
-
- Knopf F., Hammond C., Chekuru A., Kurth T., Hans S., Weber C.W., Mahatma G., Fisher S., Brand M., Schulte-Merker S., Weidinger G. Bone Regenerates via Dedifferentiation of Osteoblasts in the Zebrafish Fin. Dev. Cell. 2011;20:713–724. - PubMed
-
- Sousa S., Afonso N., Bensimon-Brito A., Fonseca M., Simões M., Leon J., Roehl H., Cancela M.L., Jacinto A., Amsterdam A., et al. Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development. 2011;138:3897–3905. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

