Targeting TACC3 Induces Immunogenic Cell Death and Enhances T-DM1 Response in HER2-Positive Breast Cancer
- PMID: 38319231
- PMCID: PMC11063689
- DOI: 10.1158/0008-5472.CAN-23-2812
Targeting TACC3 Induces Immunogenic Cell Death and Enhances T-DM1 Response in HER2-Positive Breast Cancer
Abstract
Trastuzumab emtansine (T-DM1) was the first and one of the most successful antibody-drug conjugates (ADC) approved for treating refractory HER2-positive breast cancer. Despite its initial clinical efficacy, resistance is unfortunately common, necessitating approaches to improve response. Here, we found that in sensitive cells, T-DM1 induced spindle assembly checkpoint (SAC)-dependent immunogenic cell death (ICD), an immune-priming form of cell death. The payload of T-DM1 mediated ICD by inducing eIF2α phosphorylation, surface exposure of calreticulin, ATP and HMGB1 release, and secretion of ICD-related cytokines, all of which were lost in resistance. Accordingly, ICD-related gene signatures in pretreatment samples correlated with clinical response to T-DM1-containing therapy, and increased infiltration of antitumor CD8+ T cells in posttreatment samples was correlated with better T-DM1 response. Transforming acidic coiled-coil containing 3 (TACC3) was overexpressed in T-DM1-resistant cells, and T-DM1 responsive patients had reduced TACC3 protein expression whereas nonresponders exhibited increased TACC3 expression during T-DM1 treatment. Notably, genetic or pharmacologic inhibition of TACC3 restored T-DM1-induced SAC activation and induction of ICD markers in vitro. Finally, TACC3 inhibition in vivo elicited ICD in a vaccination assay and potentiated the antitumor efficacy of T-DM1 by inducing dendritic cell maturation and enhancing intratumoral infiltration of cytotoxic T cells. Together, these results illustrate that ICD is a key mechanism of action of T-DM1 that is lost in resistance and that targeting TACC3 can restore T-DM1-mediated ICD and overcome resistance.
Significance: Loss of induction of immunogenic cell death in response to T-DM1 leads to resistance that can be overcome by targeting TACC3, providing an attractive strategy to improve the efficacy of T-DM1.
©2024 The Authors; Published by the American Association for Cancer Research.
Figures
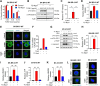
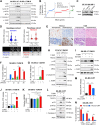
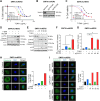
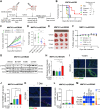
![Figure 1. T-DM1 induces ICD markers in T-DM1–sensitive breast cancer cells and ICD correlates with T-DM1 sensitivity in patients. A, Western blot analysis of mitotic arrest [p-H3 (S10)], apoptosis (cleaved caspase-3 and PARP), and ICD marker [p-eIF2α (S51)] in T-DM1–treated SK-BR-3 WT (left) and BT-474 WT (right) cells. B and C, Relative ATP release (B) and HMGB1 release (C) from T-DM1–treated SK-BR-3 WT and BT-474 WT cells (n = 3, 4). D, Immunofluorescence cell-surface staining of calreticulin (green) in T-DM1–treated SK-BR-3 WT cells. Scale bar, 10 μm. DAPI was used to stain the nucleus. Its quantification is provided on the right. E, Cytokine array blot analysis showing the differentially secreted cytokines in T-DM1–treated SK-BR-3 WT cells. F, Schematic summary of the treatment scheme and the sample collection timeline in GSE194040 (22). G, Heatmap of ICD-related genes found in the ICD gene signature score (33) and their correlation with pCR in T-DM1 + pertuzumab-treated patients from GSE194040. pCR: 1, sensitive; pCR: 0, resistant. Chi-square analysis of sensitive vs. resistant tumors expressing low vs. high ICD score is provided below. H, Percentage of CD8+ cells in sensitive (sens) vs. resistant (res) tumors collected pre- (n = 40) and post-T-DM1 (n = 18) treatment. Tables of the percentages of CD8-low or CD8-high tumors (based on average levels of CD8+ cells in each group) are given below and significance was calculated using Chi-square test. I, The representative images from H. Scale bar, 150 μm. Data correspond to mean values ± SD. P values for the bar graphs were calculated with the unpaired, two-tailed Student t test. Significance for the Chi-square analysis was calculated with the Chi-square test. **, P < 0.01. (F, Created with BioRender.com.)](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/e987/11063689/06be0edb16cc/1475fig1.gif)

Update of
-
Reviving immunogenic cell death upon targeting TACC3 enhances T-DM1 response in HER2-positive breast cancer.bioRxiv [Preprint]. 2023 Sep 14:2023.09.12.557273. doi: 10.1101/2023.09.12.557273. bioRxiv. 2023. Update in: Cancer Res. 2024 May 2;84(9):1475-1490. doi: 10.1158/0008-5472.CAN-23-2812. PMID: 37745348 Free PMC article. Updated. Preprint.
References
-
- Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol 2017;17:97–111. - PubMed
-
- Kroemer G, Galassi C, Zitvogel L, Galluzzi L. Immunogenic cell stress and death. Nat Immunol 2022;23:487–500. - PubMed
-
- Galluzzi L, Humeau J, Buque A, Zitvogel L, Kroemer G. Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nat Rev Clin Oncol 2020;17:725–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

