Molecular pathological classification of colorectal cancer-an update
- PMID: 38319359
- PMCID: PMC10948573
- DOI: 10.1007/s00428-024-03746-3
Molecular pathological classification of colorectal cancer-an update
Erratum in
-
Correction to: Molecular pathological classification of colorectal cancer-an update.Virchows Arch. 2024 Feb;484(2):287. doi: 10.1007/s00428-024-03773-0. Virchows Arch. 2024. PMID: 38416216 Free PMC article. No abstract available.
Abstract
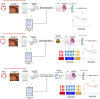
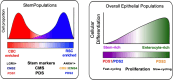
Colorectal cancer (CRC) has a broad range of molecular alterations with two major mechanisms of genomic instability (chromosomal instability and microsatellite instability) and has been subclassified into 4 consensus molecular subtypes (CMS) based on bulk RNA sequence data. Here, we update the molecular pathological classification of CRC with an overview of more recent bulk and single-cell RNA data analysis for development of transcriptional classifiers and risk stratification methods, taking into account the marked inter-tumoural and intra-tumoural heterogeneity of CRC. The importance of the stromal and immune components or tumour microenvironment (TME) to prognosis has emerged from these analyses. Attempts to remove the contribution of the tumour microenvironment and reveal neoplastic-specific transcriptional traits involved identification of the CRC intrinsic subtypes (CRIS). The use of immunohistochemistry and digital pathology to implement classification systems are evolving fields. Conventional adenoma versus serrated polyp pathway transcriptomic analysis and characterisation of canonical LGR5+ crypt base columnar stem cell versus ANXA1+ regenerative stem cell phenotypes emerged as key properties for improved understanding of transcriptional signals involved in molecular subclassification of colorectal cancers. Recently, classification by three pathway-derived subtypes (PDS1-3) has been developed, revealing a continuum of intrinsic biology associated with biological, stem cell, histopathological, and clinical attributes.
Keywords: Colorectal cancer; Hereditary; Molecular pathological classification.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

