Adipocyte-Specific Hnrnpa1 Knockout Aggravates Obesity-Induced Metabolic Dysfunction via Upregulation of CCL2
- PMID: 38320300
- PMCID: PMC11043064
- DOI: 10.2337/db23-0609
Adipocyte-Specific Hnrnpa1 Knockout Aggravates Obesity-Induced Metabolic Dysfunction via Upregulation of CCL2
Abstract
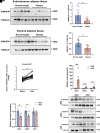
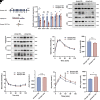
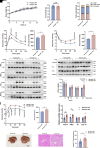
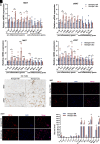
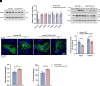
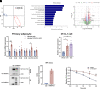
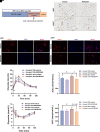
Heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1) is involved in lipid and glucose metabolism via mRNA processing. However, whether and how HNRNPA1 alters adipocyte function in obesity remain obscure. Here, we found that the obese state downregulated HNRNPA1 expression in white adipose tissue (WAT). The depletion of adipocyte HNRNPA1 promoted markedly increased macrophage infiltration and expression of proinflammatory and fibrosis genes in WAT of obese mice, eventually leading to exacerbated insulin sensitivity, glucose tolerance, and hepatic steatosis. Mechanistically, HNRNPA1 interacted with Ccl2 and regulated its mRNA stability. Intraperitoneal injection of CCL2-CCR2 signaling antagonist improved adipose tissue inflammation and systemic glucose homeostasis. Furthermore, HNRNPA1 expression in human WAT was negatively correlated with BMI, fat percentage, and subcutaneous fat area. Among individuals with 1-year metabolic surgery follow-up, HNRNPA1 expression was positively related to percentage of total weight loss. These findings identify adipocyte HNRNPA1 as a link between adipose tissue inflammation and systemic metabolic homeostasis, which might be a promising therapeutic target for obesity-related disorders.
© 2024 by the American Diabetes Association.
Conflict of interest statement
Figures

Similar articles
-
Siah2 modulates sex-dependent metabolic and inflammatory responses in adipose tissue to a high-fat diet challenge.Biol Sex Differ. 2019 Apr 15;10(1):19. doi: 10.1186/s13293-019-0233-y. Biol Sex Differ. 2019. PMID: 30987673 Free PMC article.
-
Adipocyte Expression of SLC19A1 Links DNA Hypermethylation to Adipose Tissue Inflammation and Insulin Resistance.J Clin Endocrinol Metab. 2018 Feb 1;103(2):710-721. doi: 10.1210/jc.2017-01382. J Clin Endocrinol Metab. 2018. PMID: 29121255
-
Specific loss of adipocyte CD248 improves metabolic health via reduced white adipose tissue hypoxia, fibrosis and inflammation.EBioMedicine. 2019 Jun;44:489-501. doi: 10.1016/j.ebiom.2019.05.057. Epub 2019 Jun 17. EBioMedicine. 2019. PMID: 31221584 Free PMC article.
-
Obesity-induced inflammation in white adipose tissue is attenuated by loss of melanocortin-3 receptor signaling.Endocrinology. 2007 Dec;148(12):6186-94. doi: 10.1210/en.2007-0699. Epub 2007 Sep 27. Endocrinology. 2007. PMID: 17901224
-
Omega-3 fatty acids and adipose tissue biology.Mol Aspects Med. 2018 Dec;64:147-160. doi: 10.1016/j.mam.2018.01.004. Epub 2018 Jan 17. Mol Aspects Med. 2018. PMID: 29329795 Review.
Cited by
-
Molecular Pathways Linking High-Fat Diet and PM2.5 Exposure to Metabolically Abnormal Obesity: A Systematic Review and Meta-Analysis.Biomolecules. 2024 Dec 16;14(12):1607. doi: 10.3390/biom14121607. Biomolecules. 2024. PMID: 39766314 Free PMC article.
-
Role of the E26 transformation specific transcription factor family in metabolic disorders.J Endocrinol Invest. 2025 Jul 5. doi: 10.1007/s40618-025-02634-0. Online ahead of print. J Endocrinol Invest. 2025. PMID: 40616738 Review.
-
Research progress on the cross-regulation between ferroptosis and immunogenic cell death in tumor micro-environment.Front Oncol. 2025 Jun 4;15:1581951. doi: 10.3389/fonc.2025.1581951. eCollection 2025. Front Oncol. 2025. PMID: 40535125 Free PMC article. Review.
-
SRSF3 and hnRNP A1-mediated m6A-modified circCDK14 regulates intramuscular fat deposition by acting as miR-4492-z sponge.Cell Mol Biol Lett. 2025 Mar 4;30(1):26. doi: 10.1186/s11658-025-00699-6. Cell Mol Biol Lett. 2025. PMID: 40038607 Free PMC article.
-
Cross-regulation between adipose tissue innervation and metaflammation: a potential therapeutic target for obesity.Am J Transl Res. 2025 Jun 15;17(6):4087-4100. doi: 10.62347/AIWS5429. eCollection 2025. Am J Transl Res. 2025. PMID: 40672614 Free PMC article. Review.
References
-
- Blüher M. Obesity: global epidemiology and pathogenesis. Nat Rev Endocrinol 2019;15:288–298 - PubMed
-
- Chavakis T, Alexaki VI, Ferrante AW Jr. Macrophage function in adipose tissue homeostasis and metabolic inflammation. Nat Immunol 2023;24:757–766 - PubMed
-
- Scheja L, Heeren J.. The endocrine function of adipose tissues in health and cardiometabolic disease. Nat Rev Endocrinol 2019;15:507–524 - PubMed
-
- Fasshauer M, Blüher M.. Adipokines in health and disease. Trends Pharmacol Sci 2015;36:461–470 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

