Direct reprogramming of non-limb fibroblasts to cells with properties of limb progenitors
- PMID: 38320485
- PMCID: PMC10932627
- DOI: 10.1016/j.devcel.2023.12.010
Direct reprogramming of non-limb fibroblasts to cells with properties of limb progenitors
Abstract
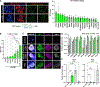
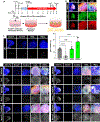
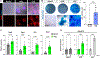
The early limb bud consists of mesenchymal limb progenitors derived from the lateral plate mesoderm (LPM). The LPM also gives rise to the mesodermal components of the flank and neck. However, the cells at these other levels cannot produce the variety of cell types found in the limb. Taking advantage of a direct reprogramming approach, we find a set of factors (Prdm16, Zbtb16, and Lin28a) normally expressed in the early limb bud and capable of imparting limb progenitor-like properties to mouse non-limb fibroblasts. The reprogrammed cells show similar gene expression profiles and can differentiate into similar cell types as endogenous limb progenitors. The further addition of Lin41 potentiates the proliferation of the reprogrammed cells. These results suggest that these same four factors may play pivotal roles in the specification of endogenous limb progenitors.
Keywords: Lin28a; Lin41; Pdrm16; Zbtb16; lateral plate mesoderm; limb bud; limb culture; limb progenitors; reprogramming.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

