A synthetic differentiation circuit in Escherichia coli for suppressing mutant takeover
- PMID: 38320549
- PMCID: PMC10882425
- DOI: 10.1016/j.cell.2024.01.024
A synthetic differentiation circuit in Escherichia coli for suppressing mutant takeover
Abstract
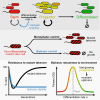
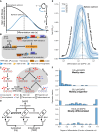
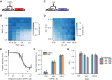
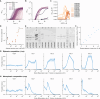
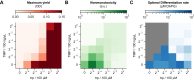
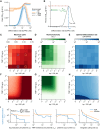
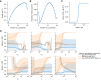
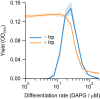
Differentiation is crucial for multicellularity. However, it is inherently susceptible to mutant cells that fail to differentiate. These mutants outcompete normal cells by excessive self-renewal. It remains unclear what mechanisms can resist such mutant expansion. Here, we demonstrate a solution by engineering a synthetic differentiation circuit in Escherichia coli that selects against these mutants via a biphasic fitness strategy. The circuit provides tunable production of synthetic analogs of stem, progenitor, and differentiated cells. It resists mutations by coupling differentiation to the production of an essential enzyme, thereby disadvantaging non-differentiating mutants. The circuit selected for and maintained a positive differentiation rate in long-term evolution. Surprisingly, this rate remained constant across vast changes in growth conditions. We found that transit-amplifying cells (fast-growing progenitors) underlie this environmental robustness. Our results provide insight into the stability of differentiation and demonstrate a powerful method for engineering evolutionarily stable multicellular consortia.
Keywords: differentiation; fitness landscape engineering; multicellular consortia; progenitors; robustness; stem cell niche; stem cells; synthetic biology; synthetic development; synthetic multicellularity; transit-amplifying cells.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Similar articles
-
SYMBIOSIS: synthetic manipulable biobricks via orthogonal serine integrase systems.Nucleic Acids Res. 2022 Mar 21;50(5):2973-2985. doi: 10.1093/nar/gkac124. Nucleic Acids Res. 2022. PMID: 35191490 Free PMC article.
-
A Synthetic Bacterial Cell-Cell Adhesion Toolbox for Programming Multicellular Morphologies and Patterns.Cell. 2018 Jul 26;174(3):649-658.e16. doi: 10.1016/j.cell.2018.06.041. Epub 2018 Jul 19. Cell. 2018. PMID: 30033369
-
Exometabolomics Assisted Design and Validation of Synthetic Obligate Mutualism.ACS Synth Biol. 2016 Jul 15;5(7):569-76. doi: 10.1021/acssynbio.5b00236. Epub 2016 Feb 17. ACS Synth Biol. 2016. PMID: 26885935
-
Overloaded and stressed: whole-cell considerations for bacterial synthetic biology.Curr Opin Microbiol. 2016 Oct;33:123-130. doi: 10.1016/j.mib.2016.07.009. Epub 2016 Aug 2. Curr Opin Microbiol. 2016. PMID: 27494248 Review.
-
Synthetic microbial consortia: from systematic analysis to construction and applications.Chem Soc Rev. 2014;43(20):6954-81. doi: 10.1039/c4cs00114a. Epub 2014 Jul 14. Chem Soc Rev. 2014. PMID: 25017039 Review.
Cited by
-
LowTempGAL: a highly responsive low temperature-inducible GAL system in Saccharomyces cerevisiae.Nucleic Acids Res. 2024 Jul 8;52(12):7367-7383. doi: 10.1093/nar/gkae460. Nucleic Acids Res. 2024. PMID: 38808673 Free PMC article.
-
Tuning and functionalization of logic gates for time resolved programming of bacterial populations.Nucleic Acids Res. 2025 Jan 7;53(1):gkae1158. doi: 10.1093/nar/gkae1158. Nucleic Acids Res. 2025. PMID: 39657755 Free PMC article.
-
Preventing Multimer Formation in Commonly Used Synthetic Biology Plasmids.ACS Synth Biol. 2025 Apr 18;14(4):1309-1315. doi: 10.1021/acssynbio.4c00508. Epub 2025 Mar 18. ACS Synth Biol. 2025. PMID: 40101192 Free PMC article.
-
Transcription regulation by RNA-induced structural strain in duplex DNA.Nucleic Acids Res. 2025 May 22;53(10):gkaf429. doi: 10.1093/nar/gkaf429. Nucleic Acids Res. 2025. PMID: 40401557 Free PMC article.
References
-
- Dao D.N., Kessin R.H., Ennis H.L. Developmental cheating and the evolutionary biology of Dictyostelium and Myxococcus. Microbiology (Reading) 2000;146:1505–1512. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

