Combining single-molecule and expansion microscopy in fission yeast to visualize protein structures at the nanostructural level
- PMID: 38320620
- PMCID: PMC10846934
- DOI: 10.1098/rsob.230414
Combining single-molecule and expansion microscopy in fission yeast to visualize protein structures at the nanostructural level
Abstract
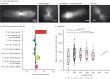
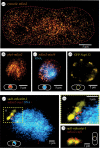
In this work, we have developed an expansion microscopy (ExM) protocol that combines ExM with photoactivated localization microscopy (ExPALM) for yeast cell imaging, and report a robust protocol for single-molecule and expansion microscopy of fission yeast, abbreviated as SExY. Our optimized SExY protocol retains about 50% of the fluorescent protein signal, doubling the amount obtained compared to the original protein retention ExM (proExM) protocol. It allows for a fivefold, highly isotropic expansion of fission yeast cells, which we carefully controlled while optimizing protein yield. We demonstrate the SExY method on several exemplary molecular targets and explicitly introduce low-abundant protein targets (e.g. nuclear proteins such as cbp1 and mis16, and the centromere-specific histone protein cnp1). The SExY protocol optimizations increasing protein yield could be beneficial for many studies, when targeting low abundance proteins, or for studies that rely on genetic labelling for various reasons (e.g. for proteins that cannot be easily targeted by extrinsic staining or in case artefacts introduced by unspecific staining interfere with data quality).
Keywords: Schizosaccharomyces pombe; correlative expansion microscopy; expansion microscopy; photoactivated localization microscopy; protein retention yield; single-molecule localization microscopy.
Conflict of interest statement
Authors declare no competing interests.
Figures

References
-
- Abbe E. 1873. Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung. Archiv f. mikrosk. Anatomie 9, 413-468. ( 10.1007/BF02956173) - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
