Epigenomic states contribute to coordinated allelic transcriptional bursting in iPSC reprogramming
- PMID: 38320809
- PMCID: PMC10847334
- DOI: 10.26508/lsa.202302337
Epigenomic states contribute to coordinated allelic transcriptional bursting in iPSC reprogramming
Abstract
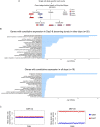
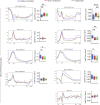
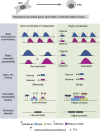
Two alleles of a gene can be transcribed independently or coordinatedly, which can lead to temporal expression heterogeneity with potentially distinct impacts on cell fate. Here, we profiled genome-wide allelic transcriptional burst kinetics during the reprogramming of MEF to induced pluripotent stem cells. We show that the degree of coordination of allelic bursting differs among genes, and alleles of many reprogramming-related genes burst in a highly coordinated fashion. Notably, we show that the chromatin accessibility of the two alleles of highly coordinated genes is similar, unlike the semi-coordinated or independent genes, suggesting the degree of coordination of allelic bursting is linked to allelic chromatin accessibility. Consistently, we show that many transcription factors have differential binding affinity between alleles of semi-coordinated or independent genes. We show that highly coordinated genes are enriched with chromatin accessibility regulators such as H3K4me3, H3K4me1, H3K36me3, H3K27ac, histone variant H3.3, and BRD4. Finally, we demonstrate that enhancer elements are highly enriched in highly coordinated genes. Our study demonstrates that epigenomic states contribute to coordinated allelic bursting to fine-tune gene expression during induced pluripotent stem cell reprogramming.
© 2024 Ayyamperumal et al.
Conflict of interest statement
S Gayen is an advisor to Bangalore Bio Cluster.
Figures










References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
