A clinically applicable connectivity signature for glioblastoma includes the tumor network driver CHI3L1
- PMID: 38320988
- PMCID: PMC10847113
- DOI: 10.1038/s41467-024-45067-8
A clinically applicable connectivity signature for glioblastoma includes the tumor network driver CHI3L1
Abstract
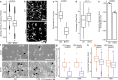
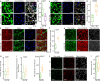
Tumor microtubes (TMs) connect glioma cells to a network with considerable relevance for tumor progression and therapy resistance. However, the determination of TM-interconnectivity in individual tumors is challenging and the impact on patient survival unresolved. Here, we establish a connectivity signature from single-cell RNA-sequenced (scRNA-Seq) xenografted primary glioblastoma (GB) cells using a dye uptake methodology, and validate it with recording of cellular calcium epochs and clinical correlations. Astrocyte-like and mesenchymal-like GB cells have the highest connectivity signature scores in scRNA-sequenced patient-derived xenografts and patient samples. In large GB cohorts, TM-network connectivity correlates with the mesenchymal subtype and dismal patient survival. CHI3L1 gene expression serves as a robust molecular marker of connectivity and functionally influences TM networks. The connectivity signature allows insights into brain tumor biology, provides a proof-of-principle that tumor cell TM-connectivity is relevant for patients' prognosis, and serves as a robust prognostic biomarker.
© 2024. The Author(s).
Conflict of interest statement
E.J., W.W. and F.W. report the patent (WO2017020982A1) entitled Agents for use in the treatment of glioma filed by the Ruprecht-Karls-Universität Heidelberg and Deutsches Krebsforschungszentrum Stiftung des öffentlichen Rechts. F.W. is co-founder of DC Europa Ltd (a company trading under the name Divide & Conquer) that is developing new medicines for the treatment of glioma. M-C.H. and J.H. report the patent (WO2020212537A1) entitled Circular permuted haloalkane transferase fusion molecules filed by the Max Planck Society. The remaining authors declare no competing interests.
Figures








References
MeSH terms
Substances
Grants and funding
- HIPO K25/Deutsches Krebsforschungszentrum (German Cancer Research Center)
- HIPO K32/Deutsches Krebsforschungszentrum (German Cancer Research Center)
- NCT 3.0 G840/Deutsches Krebsforschungszentrum (German Cancer Research Center)
- NCT 3.0 G840/Deutsches Krebsforschungszentrum (German Cancer Research Center)
- HIPO K32/Deutsches Krebsforschungszentrum (German Cancer Research Center)
- HIPO K32/Deutsches Krebsforschungszentrum (German Cancer Research Center)
- HIPO H057/Deutsches Krebsforschungszentrum (German Cancer Research Center)
- HIPO K25/Deutsches Krebsforschungszentrum (German Cancer Research Center)
- HIPO K32/Deutsches Krebsforschungszentrum (German Cancer Research Center)
- SFB 1389/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 1389/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 1389/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 1389/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 1389/Deutsche Forschungsgemeinschaft (German Research Foundation)
LinkOut - more resources
Full Text Sources
Medical

