Smith-specific regulatory T cells halt the progression of lupus nephritis
- PMID: 38321013
- PMCID: PMC10847119
- DOI: 10.1038/s41467-024-45056-x
Smith-specific regulatory T cells halt the progression of lupus nephritis
Abstract
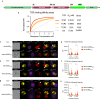
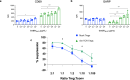
Antigen-specific regulatory T cells (Tregs) suppress pathogenic autoreactivity and are potential therapeutic candidates for autoimmune diseases such as systemic lupus erythematosus (SLE). Lupus nephritis is associated with autoreactivity to the Smith (Sm) autoantigen and the human leucocyte antigen (HLA)-DR15 haplotype; hence, we investigated the potential of Sm-specific Tregs (Sm-Tregs) to suppress disease. Here we identify a HLA-DR15 restricted immunodominant Sm T cell epitope using biophysical affinity binding assays, then identify high-affinity Sm-specific T cell receptors (TCRs) using high-throughput single-cell sequencing. Using lentiviral vectors, we transduce our lead Sm-specific TCR into Tregs derived from patients with SLE who are anti-Sm and HLA-DR15 positive. Compared with polyclonal mock-transduced Tregs, Sm-Tregs potently suppress Sm-specific pro-inflammatory responses in vitro and suppress disease progression in a humanized mouse model of lupus nephritis. These results show that Sm-Tregs are a promising therapy for SLE.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: Relevant funding was awarded by Lupus Research Alliance Inc., Funder ID 100012051, 588087, and 850279 (J.D.O.), National Health and Medical Research Council (NHMRC), Funder ID 501100000925, 2017877 (J.D.O.), Amgen Inc., Funder ID 100002429, 2021026213-001 (J.D.O.), United States Department of Defense, Funder id 100000005, LR210065 (J.D.O.). The research has been partially supported by Amgen. PCT patent application (PCT/AU2021/050254) has been filed by Monash University as the sole applicant. Inventors: J.D.O., P.J.E., and E.F.M. The remaining authors declare no competing interests.
Figures








References
MeSH terms
Substances
Grants and funding
- 100012051, 588087 and 850279/Lupus Research Alliance (Lupus Research Alliance, Inc.)
- 501100000925, 2017877/Department of Health | National Health and Medical Research Council (NHMRC)
- 100002429, 2021026213-001/Amgen (Amgen Inc.)
- 100000005, LR210065/U.S. Department of Defense (United States Department of Defense)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

