Characterization of fish-specific IFNγ-related binding with a unique receptor complex and signaling through a novel pathway
- PMID: 38321830
- PMCID: PMC10988753
- DOI: 10.1002/2211-5463.13769
Characterization of fish-specific IFNγ-related binding with a unique receptor complex and signaling through a novel pathway
Abstract
Unlike mammals, fish express two type II interferons, IFNγ and fish-specific IFNγ (IFNγ-related or IFNγrel). We previously reported the presence of two IFNγrel genes, IFNγrel 1 and IFNγrel 2, which exhibit potent antiviral activity in the Ginbuna crucian carp, Carassius auratus langsdorfii. We also found that IFNγrel 1 increased allograft rejection; however, the IFNγrel 1 receptor(s) and signaling pathways underlying this process have not yet been elucidated. In this study, we examined the unique signaling mechanism of IFNγrel 1 and its receptors. The phosphorylation and transcriptional activation of STAT6 in response to recombinant Ginbuna IFNγrel 1 (rgIFNγrel 1) was observed in Ginbuna-derived cells. Binding of rgIFNγrel 1 to Class II cytokine receptor family members (Crfbs), Crfb5 and Crfb17, which are also known as IFNAR1 and IFNGR1-1, respectively, was detected by flow cytometry. Expression of the IFNγrel 1-inducible antiviral gene, Isg15, was highest in Crfb5- and Crfb17-overexpressing GTS9 cells. Dimerization of Crfb5 and Crfb17 was detected by chemical crosslinking. The results indicate that IFNγrel 1 activates Stat6 through an interaction with unique pairs of receptors, Crfb5 and Crfb17. Indeed, this cascade is distinct from not only that of IFNγ but also that of known IFNs in other vertebrates. IFNs may be classified by their receptor and signal transduction pathways. Taken together, IFNγrel 1 may be classified as a novel type of IFN family member in vertebrates. Our findings provide important information on interferon gene evolution in bony fish.
Keywords: IFNγ‐related; STAT; cytokine receptor; signal transduction; type II interferon.
© 2024 The Authors. FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


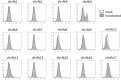
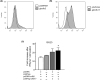


References
-
- Samuel CE (2007) Interferons, interferon receptors, signal transducer and transcriptional activators, and interferon regulatory factors. J Biol Chem 282, 20045–20046.
-
- Lutfalla G, Roest Crollius H, Stange‐Thomann N, Jaillon O, Mogensen K and Monneron D (2003) Comparative genomic analysis reveals independent expansion of a lineage‐specific gene family in vertebrates: the class II cytokine receptors and their ligands in mammals and fish. BMC Genomics 4, 29. - PMC - PubMed
-
- Krause CD and Pestka S (2005) Evolution of the class 2 cytokines and receptors, and discovery of new friends and relatives. Pharmacol Ther 106, 299–346. - PubMed
-
- Levraud JP, Boudinot P, Colin I, Benmansour A, Peyrieras N, Herbomel P and Lutfalla G (2007) Identification of the zebrafish IFN receptor: implications for the origin of the vertebrate IFN system. J Immunol 178, 4385–4394. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

