Tankyrase-1 regulates RBP-mediated mRNA turnover to promote muscle fiber formation
- PMID: 38321934
- PMCID: PMC11040007
- DOI: 10.1093/nar/gkae059
Tankyrase-1 regulates RBP-mediated mRNA turnover to promote muscle fiber formation
Abstract
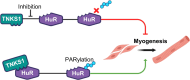
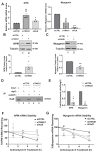
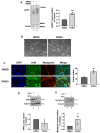
Poly(ADP-ribosylation) (PARylation) is a post-translational modification mediated by a subset of ADP-ribosyl transferases (ARTs). Although PARylation-inhibition based therapies are considered as an avenue to combat debilitating diseases such as cancer and myopathies, the role of this modification in physiological processes such as cell differentiation remains unclear. Here, we show that Tankyrase1 (TNKS1), a PARylating ART, plays a major role in myogenesis, a vital process known to drive muscle fiber formation and regeneration. Although all bona fide PARPs are expressed in muscle cells, experiments using siRNA-mediated knockdown or pharmacological inhibition show that TNKS1 is the enzyme responsible of catalyzing PARylation during myogenesis. Via this activity, TNKS1 controls the turnover of mRNAs encoding myogenic regulatory factors such as nucleophosmin (NPM) and myogenin. TNKS1 mediates these effects by targeting RNA-binding proteins such as Human Antigen R (HuR). HuR harbors a conserved TNKS-binding motif (TBM), the mutation of which not only prevents the association of HuR with TNKS1 and its PARylation, but also precludes HuR from regulating the turnover of NPM and myogenin mRNAs as well as from promoting myogenesis. Therefore, our data uncover a new role for TNKS1 as a key modulator of RBP-mediated post-transcriptional events required for vital processes such as myogenesis.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






References
-
- Ubersax J.A., Ferrell J.E. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007; 8:530–541. - PubMed
-
- Choudhary C., Weinert B.T., Nishida Y., Verdin E., Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014; 15:536–550. - PubMed
-
- Rape M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 2018; 19:59–70. - PubMed
-
- Seeler J.S., Dejean A. SUMO and the robustness of cancer. Nat. Rev. Cancer. 2017; 17:184–197. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

