The origin of different bending stiffness between double-stranded RNA and DNA revealed by magnetic tweezers and simulations
- PMID: 38321947
- PMCID: PMC10954459
- DOI: 10.1093/nar/gkae063
The origin of different bending stiffness between double-stranded RNA and DNA revealed by magnetic tweezers and simulations
Abstract
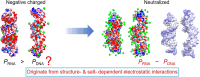
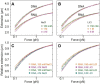
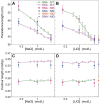
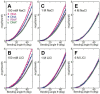
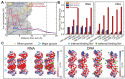
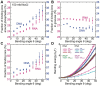
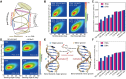
The subtle differences in the chemical structures of double-stranded (ds) RNA and DNA lead to significant variations in their biological roles and medical implications, largely due to their distinct biophysical properties, such as bending stiffness. Although it is well known that A-form dsRNA is stiffer than B-form dsDNA under physiological salt conditions, the underlying cause of this difference remains unclear. In this study, we employ high-precision magnetic-tweezer experiments along with molecular dynamics simulations and reveal that the relative bending stiffness between dsRNA and dsDNA is primarily determined by the structure- and salt-concentration-dependent ion distribution around their helical structures. At near-physiological salt conditions, dsRNA shows a sparser ion distribution surrounding its phosphate groups compared to dsDNA, causing its greater stiffness. However, at very high monovalent salt concentrations, phosphate groups in both dsRNA and dsDNA become fully neutralized by excess ions, resulting in a similar intrinsic bending persistence length of approximately 39 nm. This similarity in intrinsic bending stiffness of dsRNA and dsDNA is coupled to the analogous fluctuations in their total groove widths and further coupled to the similar fluctuation of base-pair inclination, despite their distinct A-form and B-form helical structures.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Effect of ethanol on the elasticities of double-stranded RNA and DNA revealed by magnetic tweezers and simulations.J Chem Phys. 2024 Aug 21;161(7):075101. doi: 10.1063/5.0211869. J Chem Phys. 2024. PMID: 39145565
-
Explaining the striking difference in twist-stretch coupling between DNA and RNA: A comparative molecular dynamics analysis.Nucleic Acids Res. 2015 Dec 2;43(21):10143-56. doi: 10.1093/nar/gkv1028. Epub 2015 Oct 12. Nucleic Acids Res. 2015. PMID: 26464435 Free PMC article.
-
Understanding the Relative Flexibility of RNA and DNA Duplexes: Stretching and Twist-Stretch Coupling.Biophys J. 2017 Mar 28;112(6):1094-1104. doi: 10.1016/j.bpj.2017.02.022. Biophys J. 2017. PMID: 28355538 Free PMC article.
-
Mechanisms and applications of peptide nucleic acids selectively binding to double-stranded RNA.Biopolymers. 2022 Feb;113(2):e23476. doi: 10.1002/bip.23476. Epub 2021 Sep 28. Biopolymers. 2022. PMID: 34581432 Review.
-
The left-handed double helical nucleic acids.Acta Biochim Pol. 2001;48(2):295-312. Acta Biochim Pol. 2001. PMID: 11732602 Review.
Cited by
-
Counterintuitive DNA destabilization by monovalent salt at high concentrations due to overcharging.Nat Commun. 2025 Jan 2;16(1):113. doi: 10.1038/s41467-024-55404-6. Nat Commun. 2025. PMID: 39747043 Free PMC article.
-
Binding of a Tricationic meso-Substituted Porphyrin to poly(A)⋅poly(U): an Experimental Study.J Fluoresc. 2024 Oct 28. doi: 10.1007/s10895-024-04000-4. Online ahead of print. J Fluoresc. 2024. PMID: 39465484
References
-
- Zhao X., Guo S., Lu C., Chen J., Le S., Fu H., Yan J. Single-molecule manipulation quantification of site-specific DNA binding. Curr. Opin. Chem. Biol. 2019; 53:106–117. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

