Mapping the epitope of PD-L1 to the paratope of the antibody durvalumab using molecular dynamics simulation
- PMID: 38322578
- PMCID: PMC10839400
- DOI: 10.62347/ISDD4626
Mapping the epitope of PD-L1 to the paratope of the antibody durvalumab using molecular dynamics simulation
Abstract
Objectives: Durvalumab, a human monoclonal antibody that stops PD-L1 from attaching itself to CD80 and PD-1, was approved by the Food and Drug Administration for use in cancer therapy. An essential stage in antibody optimization is mapping paratope residues to epitope residues. In this study, our earlier computer-aided method based on molecular dynamics (MD) simulations was used to observe the paratope residues on durvalumab and their companions on PD-L1.
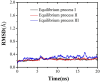
Methods: The durvalumab/PD-L1 complex model was obtained from the Protein Data Bank and used in a rectangular box for solvation. On durvalumab, the paratope residues and their companions on PD-L1 were identified using MD simulations. The interface residues were ranked on the basis of their contributions to the binding of durvalumab and PD-L1 by assessing the stability of hydrogen bonds and salt bridges. This assessment was conducted using free and guided MD simulations.
Results: Seventeen residues, including ASP26, GLU58, GLU60, ASP61, ARG113, ARG125, and THR127 on PD-L1 and H31ARG, H52LYS, H53GLN, H57GLU, H99GLU, H103PHE, H113ARG, L28ARG, L31SER, and L92TYR on durvalumab, were expected to be necessary for the binding of durvalumab to PD-L1. ASP26, ARG113, and ARG125 on PD-L1 were essential for its binding to PD-1. Eight residues (GLU60, ASP61, and THR127 on PD-L1 and L31SER, H99GLU, H53GLU, H31ARG, and H113ARG on durvalumab) were newly found, and two residues (LYS124 on PD-L1 and L94SER on durvalumab) proven nonessential for complexation, compared to the findings from the examined crystal structure.
Conclusions: The antithrombotic antibody of durvalumab's paratope may be effectively mapped to the PD-L1 epitope using the existing computer method. This information will help optimize durvalumab.
Keywords: Durvalumab; MD simulations; PD-L1; epitope; paratope.
AJTR Copyright © 2024.
Conflict of interest statement
None.
Figures




Similar articles
-
Computer prediction of paratope on antithrombotic antibody 10B12 and epitope on platelet glycoprotein VI via molecular dynamics simulation.Biomed Eng Online. 2016 Dec 28;15(Suppl 2):152. doi: 10.1186/s12938-016-0272-0. Biomed Eng Online. 2016. PMID: 28155721 Free PMC article.
-
Mapping paratope on antithrombotic antibody 6B4 to epitope on platelet glycoprotein Ibalpha via molecular dynamic simulations.PLoS One. 2012;7(7):e42263. doi: 10.1371/journal.pone.0042263. Epub 2012 Jul 30. PLoS One. 2012. PMID: 22860101 Free PMC article.
-
Safety and antitumour activity of durvalumab plus tremelimumab in non-small cell lung cancer: a multicentre, phase 1b study.Lancet Oncol. 2016 Mar;17(3):299-308. doi: 10.1016/S1470-2045(15)00544-6. Epub 2016 Feb 6. Lancet Oncol. 2016. PMID: 26858122 Free PMC article. Clinical Trial.
-
Durvalumab: an investigational anti-PD-L1 monoclonal antibody for the treatment of urothelial carcinoma.Drug Des Devel Ther. 2018 Jan 23;12:209-215. doi: 10.2147/DDDT.S141491. eCollection 2018. Drug Des Devel Ther. 2018. PMID: 29416316 Free PMC article. Review.
-
Durvalumab for the treatment of non-small cell lung cancer.Expert Rev Respir Med. 2018 Aug;12(8):627-639. doi: 10.1080/17476348.2018.1494575. Epub 2018 Jul 9. Expert Rev Respir Med. 2018. PMID: 29958099 Review.
References
-
- Callahan MK, Postow MA, Wolchok JD. Targeting T cell co-receptors for cancer therapy. Immunity. 2016;44:1069–1078. - PubMed
-
- Zak KM, Grudnik P, Magiera K, Domling A, Dubin G, Holak TA. Structural biology of the immune checkpoint receptor PD-1 and its ligands PD-L1/PD-L2. Structure. 2017;25:1163–1174. - PubMed
-
- Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, Fitz LJ, Malenkovich N, Okazaki T, Byrne MC, Horton HF, Fouser L, Carter L, Ling V, Bowman MR, Carreno BM, Collins M, Wood CR, Honjo T. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. 2000;192:1027–1034. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
